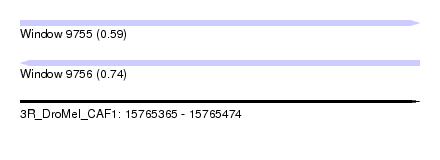
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,765,365 – 15,765,474 |
| Length | 109 |
| Max. P | 0.737714 |

| Location | 15,765,365 – 15,765,474 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.50 |
| Mean single sequence MFE | -19.82 |
| Consensus MFE | -13.68 |
| Energy contribution | -13.92 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.586432 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15765365 109 + 27905053 ACAUAGUCA-----CACUUAGCAAUCAAACUAGACAA-AAGUUUCUCGAUUAAAAUGAGCAACUU-UUGUGAAA---UUCUCUUAUCUAACAAUUUCCGCUGCUUAGUUGAA-CCGAAAU ......(((-----.(((.((((..........((((-(((((.((((.......)))).)))))-))))((((---((............))))))...)))).)))))).-....... ( -19.90) >DroSec_CAF1 22542 100 + 1 --------------CACUUAGCAAUCAAACUAGACAA-AAGUUUCUCGAUUAAAAUGAGCAACUU-UUGUGAAA---UUCUCUUAUCUAACAAUUUCCGCUGCUUAGCUGAA-CCGAAAU --------------...(((((...........((((-(((((.((((.......)))).)))))-))))((((---((............)))))).........))))).-....... ( -17.40) >DroSim_CAF1 22532 100 + 1 --------------CACUUAGCAAUCAAACUAGACAA-AAGUUUCUCGAUUAAAAUGAGCAACUU-UUGUGAAA---UUCUCUUAUCUAACAAUUUCCGCUGCUUAGUUGAA-CCGAAAU --------------.(((.((((..........((((-(((((.((((.......)))).)))))-))))((((---((............))))))...)))).)))....-....... ( -17.70) >DroEre_CAF1 24688 108 + 1 -CAUAGUCA-----CACUUAGCAAUCAAACUAGACAA-AAGUUUCUCGAUUAAAAUGAGCAACUU-UUGCGAAA---UUCUCUUAUCUAACAAUUUCCGCUGCUUAGUUGAA-CCGAAAU -.....(((-----.(((.((((.........(.(((-(((((.((((.......)))).)))))-))))((((---((............))))))...)))).)))))).-....... ( -18.90) >DroPer_CAF1 24401 119 + 1 -UAUAGUUUCUGUUCUCUGAGCAAUCAAACUAGACAAAAAGUUUCUCGAUUAAAAUGAGCAACUUUUUGUGAGAAUUUUCUCUUAUCUGACAAUUUCCGCUGCUUAGCCGAAGACCAAAU -..((((((.(((((...)))))...)))))).((((((((((.((((.......)))).))))))))))((((....))))...........((((.(((....))).))))....... ( -25.20) >consensus __AUAGU_______CACUUAGCAAUCAAACUAGACAA_AAGUUUCUCGAUUAAAAUGAGCAACUU_UUGUGAAA___UUCUCUUAUCUAACAAUUUCCGCUGCUUAGUUGAA_CCGAAAU ................((.((((..........((((((((((.((((.......)))).))))))))))(((....)))....................)))).))............. (-13.68 = -13.92 + 0.24)



| Location | 15,765,365 – 15,765,474 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.50 |
| Mean single sequence MFE | -25.06 |
| Consensus MFE | -19.86 |
| Energy contribution | -19.82 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15765365 109 - 27905053 AUUUCGG-UUCAACUAAGCAGCGGAAAUUGUUAGAUAAGAGAA---UUUCACAA-AAGUUGCUCAUUUUAAUCGAGAAACUU-UUGUCUAGUUUGAUUGCUAAGUG-----UGACUAUGU .....((-((..(((.(((((((((((((.((......)).))---))))((((-(((((.(((.........))).)))))-))))......)).))))).))).-----.)))).... ( -25.80) >DroSec_CAF1 22542 100 - 1 AUUUCGG-UUCAGCUAAGCAGCGGAAAUUGUUAGAUAAGAGAA---UUUCACAA-AAGUUGCUCAUUUUAAUCGAGAAACUU-UUGUCUAGUUUGAUUGCUAAGUG-------------- .......-....(((.(((((((((((((.((......)).))---))))((((-(((((.(((.........))).)))))-))))......)).))))).))).-------------- ( -22.90) >DroSim_CAF1 22532 100 - 1 AUUUCGG-UUCAACUAAGCAGCGGAAAUUGUUAGAUAAGAGAA---UUUCACAA-AAGUUGCUCAUUUUAAUCGAGAAACUU-UUGUCUAGUUUGAUUGCUAAGUG-------------- .......-....(((.(((((((((((((.((......)).))---))))((((-(((((.(((.........))).)))))-))))......)).))))).))).-------------- ( -22.60) >DroEre_CAF1 24688 108 - 1 AUUUCGG-UUCAACUAAGCAGCGGAAAUUGUUAGAUAAGAGAA---UUUCGCAA-AAGUUGCUCAUUUUAAUCGAGAAACUU-UUGUCUAGUUUGAUUGCUAAGUG-----UGACUAUG- .....((-((..(((.(((((((((((((.((......)).))---))))((((-(((((.(((.........))).)))))-))))......)).))))).))).-----.))))...- ( -26.10) >DroPer_CAF1 24401 119 - 1 AUUUGGUCUUCGGCUAAGCAGCGGAAAUUGUCAGAUAAGAGAAAAUUCUCACAAAAAGUUGCUCAUUUUAAUCGAGAAACUUUUUGUCUAGUUUGAUUGCUCAGAGAACAGAAACUAUA- .((((.(((((.(((....))).)))...(((((((..((((....))))((((((((((.(((.........))).))))))))))...)))))))........)).)))).......- ( -27.90) >consensus AUUUCGG_UUCAACUAAGCAGCGGAAAUUGUUAGAUAAGAGAA___UUUCACAA_AAGUUGCUCAUUUUAAUCGAGAAACUU_UUGUCUAGUUUGAUUGCUAAGUG_______ACUAU__ ............(((.(((((((((.............((((....))))((((((((((.(((.........))).))))))))))....)))).))))).)))............... (-19.86 = -19.82 + -0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:32 2006