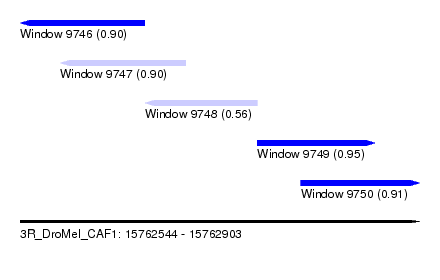
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,762,544 – 15,762,903 |
| Length | 359 |
| Max. P | 0.953141 |

| Location | 15,762,544 – 15,762,656 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.48 |
| Mean single sequence MFE | -35.60 |
| Consensus MFE | -13.50 |
| Energy contribution | -16.26 |
| Covariance contribution | 2.76 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.38 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15762544 112 - 27905053 CUU--CAGGGGCCAAGCCAGUCUUUUGUUGCACUCACUUUGUGUUGUGAUGACAUUGGCUCUGGCUCAGGAGAAGUGA---GAAUGGAUCCCAUGG---AAACAUCAUUAGAUCGCGUUA (((--(..((((((((((((((..(..(.((((.......)))).)..).)))..))))).)))))).))))..((((---.((((......(((.---...)))))))...)))).... ( -36.30) >DroSec_CAF1 19740 112 - 1 CUU--CAGGGGCCAAGCCAGUCCUUUGUUACACUCACUUUGUGUUGUGAUGACAUUGGCUCUGGCUCAGGAAUAGGAA---GAAUGGAUCCCAUGG---AAACAUCAUUAGAUCGCGUUA (((--(..(..((.((((((.((..((((((((.(((...)))..))).)))))..))..))))))..))..)..)))---)((((((((..(((.---...))).....)))).)))). ( -37.30) >DroSim_CAF1 19727 112 - 1 CUU--CAGGGGCCAAGCCAGUCUUUUGUUGCACUCACUUUGUGUUGUGAUGACAUUGGCUCUGGCUCAGGAAUAGGAA---GAAUGGAUCCCAUGG---AAACAUCAUUAGAUCGCGUUA .((--(..((((((((((((((..(..(.((((.......)))).)..).)))..))))).))))))..)))......---.((((((((..(((.---...))).....)))).)))). ( -35.90) >DroEre_CAF1 21941 92 - 1 CAU--CAGGGGCCAAGUCAGUCCUUUGUUGCACUCACUUUGAGUUGUGAUGACAUUGGUUCUG-----------------------GAUCCCAUGG---AACCAUCAUUAGACCGCCUCA ...--..(((((...(((.(((....((..(((((.....))).))..)))))..((((((((-----------------------.......)))---)))))......))).))))). ( -28.30) >DroPer_CAF1 20279 111 - 1 AUGGACGGGGCCCAGGCC----CUAUGGGUCUUUGCCUUUGUGU-----UGACAUCGGUUCAGCCAAAGGUUAGGGAAAUUGAAUGGAUCCCAUGGUGGUGCCAUCAUUAGAUCGCAUUA .....(((..((((((((----(...))))))..(((((((.((-----(((.......))))))))))))..)))...)))((((((((..((((((....))))))..)))).)))). ( -40.20) >consensus CUU__CAGGGGCCAAGCCAGUCCUUUGUUGCACUCACUUUGUGUUGUGAUGACAUUGGCUCUGGCUCAGGAAAAGGAA___GAAUGGAUCCCAUGG___AAACAUCAUUAGAUCGCGUUA ........(((((((((((((...(..(.((((.......)))).)..)....))))))).))))))...................((((..((((........))))..))))...... (-13.50 = -16.26 + 2.76)

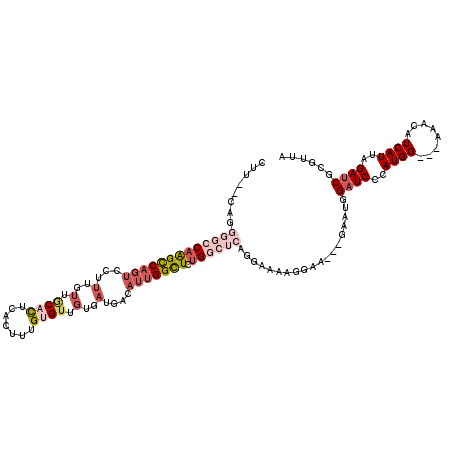
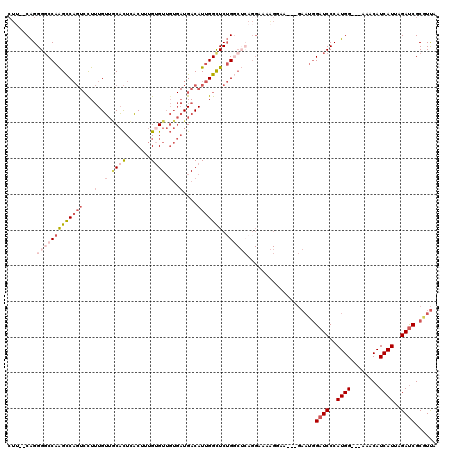
| Location | 15,762,580 – 15,762,693 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.17 |
| Mean single sequence MFE | -38.60 |
| Consensus MFE | -16.60 |
| Energy contribution | -18.16 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.43 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15762580 113 - 27905053 CGUUUUGUAUCCCAUGAGGGUGC---GAAACCCCUUUGAUCUU--CAGGGGCCAAGCCAGUCUUUUGUUGCACUCACUUUGUGUUGUGAUGACAUUGGCUCUGGCUCAGGAGAAGUGA-- .(((((((((((.....))))))---)))))........((((--(..((((((((((((((..(..(.((((.......)))).)..).)))..))))).)))))).))))).....-- ( -42.60) >DroSec_CAF1 19776 113 - 1 CGUUUUGUAUCCCAUGAGGGUGC---GAAACCCCUUUGAUCUU--CAGGGGCCAAGCCAGUCCUUUGUUACACUCACUUUGUGUUGUGAUGACAUUGGCUCUGGCUCAGGAAUAGGAA-- .(((((((((((.....))))))---))))).(((......((--(..(((((((((((((...(..(((((..(((...))).)))))..).))))))).))))))..))).)))..-- ( -42.20) >DroSim_CAF1 19763 113 - 1 CGUUUUGUAUCCCAUGAGGGUGC---GAAACCCCUUUGAUCUU--CAGGGGCCAAGCCAGUCUUUUGUUGCACUCACUUUGUGUUGUGAUGACAUUGGCUCUGGCUCAGGAAUAGGAA-- .(((((((((((.....))))))---))))).(((......((--(..((((((((((((((..(..(.((((.......)))).)..).)))..))))).))))))..))).)))..-- ( -41.70) >DroEre_CAF1 21972 98 - 1 CGUUUUGUGUGCCAUGAGGGUGC---GAAAUCCCUUGGAUCAU--CAGGGGCCAAGUCAGUCCUUUGUUGCACUCACUUUGAGUUGUGAUGACAUUGGUUCUG----------------- ......(((..(((..((((...---.....)))))))..)))--..((((((((((((........(..(((((.....))).))..))))).)))))))).----------------- ( -26.40) >DroPer_CAF1 20319 111 - 1 CCUUUUGUAUCCCAAGAGGGUGCCAGGAGAACCCUUUGACAUGGACGGGGCCCAGGCC----CUAUGGGUCUUUGCCUUUGUGU-----UGACAUCGGUUCAGCCAAAGGUUAGGGAAAU ..........(((((((((((.........))))))).........(((((....)))----)).))))((((((((((((.((-----(((.......))))))))))).))))))... ( -40.10) >consensus CGUUUUGUAUCCCAUGAGGGUGC___GAAACCCCUUUGAUCUU__CAGGGGCCAAGCCAGUCCUUUGUUGCACUCACUUUGUGUUGUGAUGACAUUGGCUCUGGCUCAGGAAAAGGAA__ .(((((((((((.....))))))...)))))((((...........))))(((((((((((...(..(.((((.......)))).)..)....))))))).))))............... (-16.60 = -18.16 + 1.56)

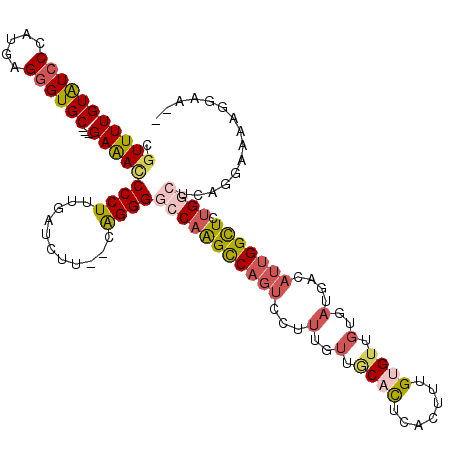
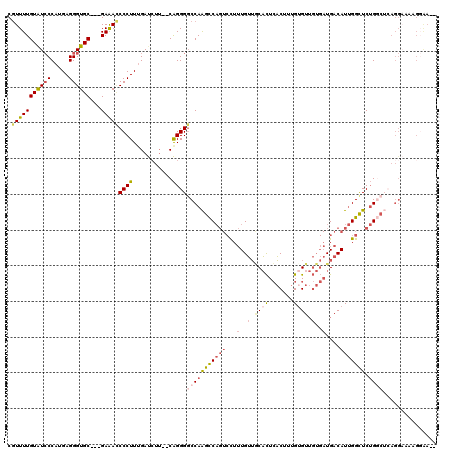
| Location | 15,762,656 – 15,762,757 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 87.98 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -24.29 |
| Energy contribution | -24.47 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15762656 101 - 27905053 GUAGCUUCAUCGGGACGUUGCUCAAAGGUCGAUGACUCAGAUCCACGUCC------UCGUUCUUUAGUAACGUUUUGUAUCCCAUGAGGGUGCGAAACCCCUUUGAU ......(((..(((..((((((.((((..(((.(((..........))).------)))..))))))))))(((((((((((.....))))))))))))))..))). ( -35.90) >DroSec_CAF1 19852 101 - 1 GUAGCUUAAUCGGGACGUAGCUCAAAGGUCGAUGACUCAGGUCCACGUCC------UCGUUUUUUAGUAUCGUUUUGUAUCCCAUGAGGGUGCGAAACCCCUUUGAU ...(((.(((.(((((((.(((....((((...))))..)))..))))))------).)))....)))...(((((((((((.....)))))))))))......... ( -27.90) >DroSim_CAF1 19839 101 - 1 GUAGCUUCAUCGGGACGUUGCUCAAAGGUCGAUGACUCAGGUCCACGUCC------UCGUUCUUUAGUAGCGUUUUGUAUCCCAUGAGGGUGCGAAACCCCUUUGAU ......(((..(((..((((((.((((..(((.(((..........))).------)))..))))))))))(((((((((((.....))))))))))))))..))). ( -35.90) >DroEre_CAF1 22033 107 - 1 AAAGCAUCGUCGGGACGUCGCCCGAAGGUCGAUGACUCUGUUCCACGUCCUCGUCCUCGUCCUUUAGUAGCGUUUUGUGUGCCAUGAGGGUGCGAAAUCCCUUGGAU ...((((...((((((((..(..(((((.(((.(((................))).))).))))).)..)))))))).))))...(((((........))))).... ( -32.29) >consensus GUAGCUUCAUCGGGACGUUGCUCAAAGGUCGAUGACUCAGGUCCACGUCC______UCGUUCUUUAGUAGCGUUUUGUAUCCCAUGAGGGUGCGAAACCCCUUUGAU ...........(((..((((((.(((((.(((.(((..........))).......))).)))))))))))(((((((((((.....))))))))))))))...... (-24.29 = -24.47 + 0.19)

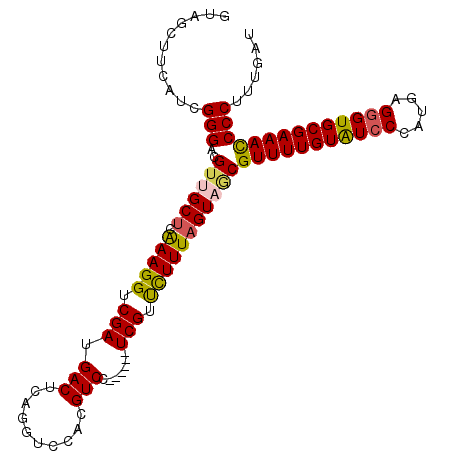
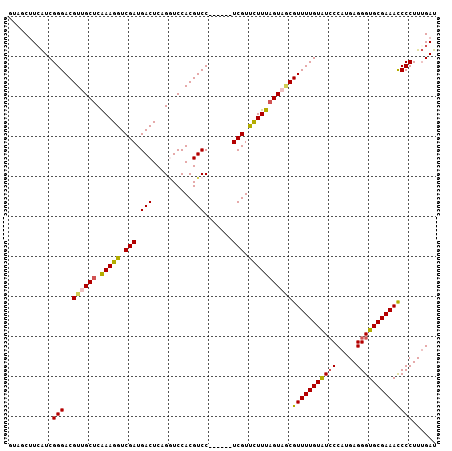
| Location | 15,762,757 – 15,762,863 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.10 |
| Mean single sequence MFE | -30.74 |
| Consensus MFE | -12.30 |
| Energy contribution | -13.42 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.40 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15762757 106 + 27905053 GAAGAAAAGGGAUCCAGGAAACCGGACCCAUGACA-UUGGAUCUGGUUUUUGCAGACCAGAUAAACUAGCUCCUACUGUAACUAA------CAAGGGG-------CACAUUGAAAGCAAC ........(((.(((.(....).))))))....((-.((.((((((((......))))))))......((((((..(((.....)------)))))))-------).)).))........ ( -33.80) >DroSec_CAF1 19953 112 + 1 GCAGGAAAG-GAUCCAGGAAACCGGACCCACGACU-UUGGAUCUGGUUUCUGACGACCAGAUAAACUAGCUCCUACUGUAACUAG------CAAGGGGUAACUAGCACACAGAAAACAAC ((.((...(-(.((..((((((((((.(((.....-.))).))))))))))...))))..........((((((..(((.....)------))))))))..)).)).............. ( -31.70) >DroSim_CAF1 19940 103 + 1 GCAGUAAAG-GAUCCAGGAAACCGGACCCACAACU-UUGGAUCUGGUUUCUGUCGACCAGAUAAACUAGCUCCUACUGUAACUAA------CAAGGGG---------CACAGAAAACAAC ........(-(.((..((((((((((.(((.....-.))).))))))))))...))))..........((((((..(((.....)------)))))))---------)............ ( -30.60) >DroEre_CAF1 22140 103 + 1 GCAGGAGGG-GCUCCAGGAAACCGGAGCCACGACU-UUCGAUCGGGUUU-UCCCGACCAGAUAAACCAGCUCCUACUGUAACUAG------CGAGGGG-------CACACGG-AAACAAC ......(((-(((((.(....).))))))......-...(.(((((...-.))))).).......)).((((((.(((....)))------..)))))-------)....(.-...)... ( -37.70) >DroPer_CAF1 20477 100 + 1 GCAGAAA-----------GAACCGGACUGAUGUUCUGCUGUUCAGGUUUC--AUGACCAGAAAAACGUGUUGUAACUACGACUACAGACUACAGACUA-------CAGAUGGAGAAGGAC .......-----------...(((..(((.((.((((..(((..((((..--..))))........(((((((....))))).)).)))..)))).))-------))).)))........ ( -19.90) >consensus GCAGAAAAG_GAUCCAGGAAACCGGACCCACGACU_UUGGAUCUGGUUUCUGACGACCAGAUAAACUAGCUCCUACUGUAACUAA______CAAGGGG_______CACACAGAAAACAAC ((((........(((.(....).)))..............((((((((......)))))))).............))))......................................... (-12.30 = -13.42 + 1.12)

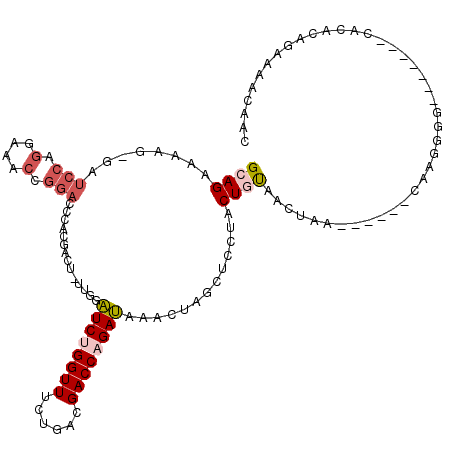
| Location | 15,762,796 – 15,762,903 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.31 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -14.31 |
| Energy contribution | -13.83 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.909361 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15762796 107 + 27905053 AUCUGGUUUUUGCAGACCAGAUAAACUAGCUCCUACUGUAACUAA------CAAGGGG-------CACAUUGAAAGCAACAUAACUAAAGCCAUGUUCUAUCGGCAUAUAAGCCAAUUAA ((((((((......))))))))......((((((..(((.....)------)))))))-------).....((.((.(((((..(....)..))))))).))(((......)))...... ( -28.00) >DroSec_CAF1 19991 114 + 1 AUCUGGUUUCUGACGACCAGAUAAACUAGCUCCUACUGUAACUAG------CAAGGGGUAACUAGCACACAGAAAACAACAUAACUAAAGCCAUGUUCUAUCGGCACAUAAGCCAAUUAA ((((((((......))))))))...((((..(((..(((.....)------))..)))...))))......((....(((((..(....)..)))))...))(((......)))...... ( -23.00) >DroSim_CAF1 19978 105 + 1 AUCUGGUUUCUGUCGACCAGAUAAACUAGCUCCUACUGUAACUAA------CAAGGGG---------CACAGAAAACAACAUAACUAAAGCCAUGUUCUAUCGGCACAUAAGCCAAUUAA ((((((((......))))))))......((((((..(((.....)------)))))))---------)...((....(((((..(....)..)))))...))(((......)))...... ( -24.60) >DroEre_CAF1 22178 105 + 1 AUCGGGUUU-UCCCGACCAGAUAAACCAGCUCCUACUGUAACUAG------CGAGGGG-------CACACGG-AAACAACAUCACUAAAGCCAUGUUCUAUCGGCACAUAAGCCAAUUAA .(((((...-.)))))...(((((((..((((((.(((....)))------..)))))-------)....(.-...).................))).))))(((......)))...... ( -27.00) >DroPer_CAF1 20506 104 + 1 UUCAGGUUUC--AUGACCAGAAAAACGUGUUGUAACUACGACUACAGACUACAGACUA-------CAGAUGGAGAAGGACAUAG-------GAUAUACUAGCGGCACAUAAGCCAAUUAU (((.((((..--..)))).)))....(((((((....))))).)).............-------(..(((........)))..-------)..........(((......)))...... ( -15.40) >consensus AUCUGGUUUCUGACGACCAGAUAAACUAGCUCCUACUGUAACUAA______CAAGGGG_______CACACAGAAAACAACAUAACUAAAGCCAUGUUCUAUCGGCACAUAAGCCAAUUAA ((((((((......)))))))).......(((((...................)))))............................................(((......)))...... (-14.31 = -13.83 + -0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:26 2006