| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,215,440 – 2,215,543 |
| Length | 103 |
| Max. P | 0.939538 |

| Location | 2,215,440 – 2,215,543 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 79.64 |
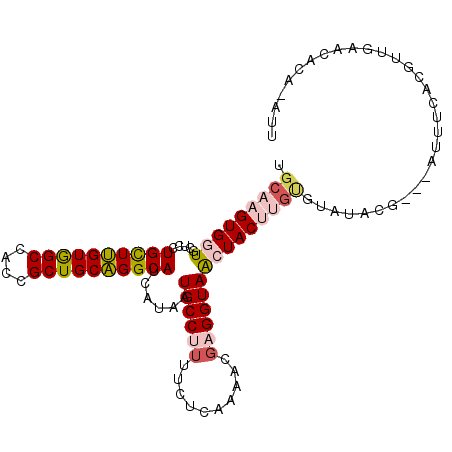
| Mean single sequence MFE | -31.07 |
| Consensus MFE | -22.21 |
| Energy contribution | -22.88 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2215440 103 - 27905053 UGCAAGUGGUGCUCCUGCUUGUGGCCACCGCUGCAGGCAUCAUAAUGCCUUUUCUCAAAACGAGGUAGCUACUUGUGAAUUUG---AUUCCACGUUGAACAUA-UUU .(((((((((.....((((((..((....))..))))))......((((((..........)))))))))))))))((((.((---.(((......))))).)-))) ( -30.40) >DroVir_CAF1 4457 97 - 1 UGCAAGUGGUGCUCCUGUUGGUAGCCACCGCUGCUGGCAUUAUAAUGCCCUUUCUCAAAACGAGGUAAGUACACGC-UAUACG---GACUUA---UGU---UAGAUA .(((.((((((((((....)).)).))))))))).(((((....)))))....(((.....)))((((((...((.-....))---.)))))---)..---...... ( -28.90) >DroSec_CAF1 4480 103 - 1 UGCAAGUGGUGCUCCUGCUUGUGGCCACCGCUGCAGGCAUCAUAAUGCCUUUUCUCAAAACUAGGUAGCUACUUGUGUAUUCG---AUUCCACGUUGAACACA-ACU .(((((((((.....((((((..((....))..))))))......(((((............))))))))))))))((.((((---((.....)))))).)).-... ( -33.00) >DroSim_CAF1 4484 103 - 1 UGCAAGUGGUGCUCCUGCUUGUGGCCACCGCUGCAGGCAUCAUAAUGCCUUUUCUCAAAACGAGGUAGCUACUUGUGUAUUCG---AUUUCACGUUGAACACA-ACU .(((((((((.....((((((..((....))..))))))......((((((..........)))))))))))))))((.((((---((.....)))))).)).-... ( -33.80) >DroYak_CAF1 3835 106 - 1 UGCAAGUGGUGCUCCUGCUUGUGGCCACCGCUGCAGGCAUCAUAAUGCCUUUUCUCAAAACGAGGUAACUACUUCUGACUACUACUAUUUCACGAUGAGCACU-AUU ....((((((((((.((((((..((....))..))))))......((((((..........)))))).............................)))))))-))) ( -31.30) >DroMoj_CAF1 4846 102 - 1 UGCAAGUGGUGCUGCUGUUGGUGGCAACCGCUGCUGGCAUUAUUAUGCCGUUUCUCAAAACGAGGUAAGUACACGG-UCUACA---UACGCA-AAUGUAUAUAUAUA .(((.(((((((..(.....)..)).)))))))).(((((....)))))....(((.....)))(((.(((((..(-(.....---...)).-..))))).)))... ( -29.00) >consensus UGCAAGUGGUGCUCCUGCUUGUGGCCACCGCUGCAGGCAUCAUAAUGCCUUUUCUCAAAACGAGGUAACUACUUGUGUAUACG___AUUUCACGUUGAACACA_AUU .(((((((((.....((((((((((....))))))))))......((((((..........)))))))))))))))............................... (-22.21 = -22.88 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:01 2006