
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,758,882 – 15,759,162 |
| Length | 280 |
| Max. P | 0.981373 |

| Location | 15,758,882 – 15,759,002 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -45.94 |
| Consensus MFE | -42.22 |
| Energy contribution | -42.34 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15758882 120 + 27905053 UGUCCUGCUUCAGCUGCACCAGCUGCUGCUGCAGUGCGUCGCUCAGCUGGAUGUAGUAGUCGAUGCCGUUGCGCAACUGCUCUUCGCAUUUCUGCAGCUUCUCGUCGCGCAUGCGCAGCA ...........(((((...)))))(((((.((((((((.((...((((((((((((((((.(.(((....)))).))))))....)))))....)))))...)).))))).)))))))). ( -45.20) >DroSec_CAF1 16078 120 + 1 UGUCCUGCUUCAGCUGCACCAGCUGCUGCUGCAGUGCGUCGCUCAGCUGGAUGUAGUAGUCGAUACCGUUGCGCAACUGCUCCUCGCAUUUCUGCAGCUUCUCGUCGCGCAUGCGCAGCA ((((..(((...(((((((((((((..((.((.....)).)).))))))).))))))))).))))..((((((((..(((.....(((....))).((........))))))))))))). ( -45.50) >DroSim_CAF1 16036 120 + 1 UGUCCUGCUUCAGCUGCACCAGCUGCUGCUGCAGUGCGUCGCUCAGCUGGAUGUAGUAGUCGAUGCCGUUGCGCAACUGCUCCUCGCAUUUCUGCAGCUUCUCGUCGCGCAUGCGCAGCA ...........(((((...)))))(((((.((((((((.((...(((((((.(((((.((((((...)))).)).))))))))..(((....)))))))...)).))))).)))))))). ( -45.40) >DroEre_CAF1 18478 120 + 1 UGUCCUGCUUCAGCUGCACCAGCUGCUGCUGCAGUGCGUCGCUCAGCUGGAUGUAGUAGUCAAUGCCGUUGCGCAACUGCUCCUCGCACUUCUGCAGCUUCUCGUCGCGCAUGCGCAGCA ...........(((((...)))))(((((.((((((((.((...(((((((.(((((.((((((...)))).)).))))))))..(((....)))))))...)).))))).)))))))). ( -45.60) >DroPer_CAF1 16220 120 + 1 UGUCCUGCUUCAGCUGGACCAGCUGCUGCUGCAGUGCGUCGCUCAGCUGGAUGUAGUAGUCGAUGCCGUUGCGCAGCUGCUCCUCGCACUUCUGCAGCUUCUCGUCGCGCUGCCGCAACA .....(((..((((..(((.((..(((((.(.((((((..(..((((((..(((((..((....))..)))))))))))..)..)))))).).)))))..)).)))..))))..)))... ( -48.00) >consensus UGUCCUGCUUCAGCUGCACCAGCUGCUGCUGCAGUGCGUCGCUCAGCUGGAUGUAGUAGUCGAUGCCGUUGCGCAACUGCUCCUCGCAUUUCUGCAGCUUCUCGUCGCGCAUGCGCAGCA ...........(((((...)))))(((((.((((((((.((...(((((((.(((((.((((((...)))).)).))))))))..(((....)))))))...)).))))).)))))))). (-42.22 = -42.34 + 0.12)



| Location | 15,758,882 – 15,759,002 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -45.38 |
| Consensus MFE | -41.30 |
| Energy contribution | -40.82 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15758882 120 - 27905053 UGCUGCGCAUGCGCGACGAGAAGCUGCAGAAAUGCGAAGAGCAGUUGCGCAACGGCAUCGACUACUACAUCCAGCUGAGCGACGCACUGCAGCAGCAGCUGGUGCAGCUGAAGCAGGACA (((..(((.((((((((.......((((....)))).......)))))))).((((...((........))..)))).)))..)))((((..((((.((....)).))))..)))).... ( -45.24) >DroSec_CAF1 16078 120 - 1 UGCUGCGCAUGCGCGACGAGAAGCUGCAGAAAUGCGAGGAGCAGUUGCGCAACGGUAUCGACUACUACAUCCAGCUGAGCGACGCACUGCAGCAGCAGCUGGUGCAGCUGAAGCAGGACA (((..(((.((((((((.......((((....)))).......)))))))).((((...((........))..)))).)))..)))((((..((((.((....)).))))..)))).... ( -43.64) >DroSim_CAF1 16036 120 - 1 UGCUGCGCAUGCGCGACGAGAAGCUGCAGAAAUGCGAGGAGCAGUUGCGCAACGGCAUCGACUACUACAUCCAGCUGAGCGACGCACUGCAGCAGCAGCUGGUGCAGCUGAAGCAGGACA (((..(((.((((((((.......((((....)))).......)))))))).((((...((........))..)))).)))..)))((((..((((.((....)).))))..)))).... ( -45.24) >DroEre_CAF1 18478 120 - 1 UGCUGCGCAUGCGCGACGAGAAGCUGCAGAAGUGCGAGGAGCAGUUGCGCAACGGCAUUGACUACUACAUCCAGCUGAGCGACGCACUGCAGCAGCAGCUGGUGCAGCUGAAGCAGGACA ..((((....((((.((.((..(((((...((((((..(..((((((.((....))...((........))))))))..)..))))))...)))))..)).)))).))....)))).... ( -43.40) >DroPer_CAF1 16220 120 - 1 UGUUGCGGCAGCGCGACGAGAAGCUGCAGAAGUGCGAGGAGCAGCUGCGCAACGGCAUCGACUACUACAUCCAGCUGAGCGACGCACUGCAGCAGCAGCUGGUCCAGCUGAAGCAGGACA ..((((..((((..(((.((..(((((...((((((..(..((((((.((....))...((........))))))))..)..))))))...)))))..)).)))..))))..)))).... ( -49.40) >consensus UGCUGCGCAUGCGCGACGAGAAGCUGCAGAAAUGCGAGGAGCAGUUGCGCAACGGCAUCGACUACUACAUCCAGCUGAGCGACGCACUGCAGCAGCAGCUGGUGCAGCUGAAGCAGGACA (((((((....)))........(((((((...((((..(..((((((.((....))...((........))))))))..)..)))))))))))..((((((...)))))).))))..... (-41.30 = -40.82 + -0.48)



| Location | 15,758,922 – 15,759,042 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -41.96 |
| Consensus MFE | -38.40 |
| Energy contribution | -38.72 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15758922 120 + 27905053 GCUCAGCUGGAUGUAGUAGUCGAUGCCGUUGCGCAACUGCUCUUCGCAUUUCUGCAGCUUCUCGUCGCGCAUGCGCAGCAUAUCAUUGGUGGUCUCCAGCAGCACCAUCACGUCCUGCUU ....(((.((((((.......((((..((((((((..(((.....(((....))).((........))))))))))))).))))...((((((((.....)).)))))))))))).))). ( -41.40) >DroSec_CAF1 16118 120 + 1 GCUCAGCUGGAUGUAGUAGUCGAUACCGUUGCGCAACUGCUCCUCGCAUUUCUGCAGCUUCUCGUCGCGCAUGCGCAGCAUAUCAUUGGUGGUCUCCAGCAGCACCAUCACGUCCUGCUU ....(((.((((((.......((((..((((((((..(((.....(((....))).((........))))))))))))).))))...((((((((.....)).)))))))))))).))). ( -41.70) >DroSim_CAF1 16076 120 + 1 GCUCAGCUGGAUGUAGUAGUCGAUGCCGUUGCGCAACUGCUCCUCGCAUUUCUGCAGCUUCUCGUCGCGCAUGCGCAGCAUAUCAUUGGUGGUCUCCAGCAGCACCAUCACGUCCUGCUU ....(((.((((((.......((((..((((((((..(((.....(((....))).((........))))))))))))).))))...((((((((.....)).)))))))))))).))). ( -41.40) >DroEre_CAF1 18518 120 + 1 GCUCAGCUGGAUGUAGUAGUCAAUGCCGUUGCGCAACUGCUCCUCGCACUUCUGCAGCUUCUCGUCGCGCAUGCGCAGCAUAUCAUUGGUGGUCUCCAGCAGCACCAUCACGUCCUGCUU ....(((.((((((.............((((((((..(((.....(((....))).((........)))))))))))))........((((((((.....)).)))))))))))).))). ( -39.30) >DroPer_CAF1 16260 120 + 1 GCUCAGCUGGAUGUAGUAGUCGAUGCCGUUGCGCAGCUGCUCCUCGCACUUCUGCAGCUUCUCGUCGCGCUGCCGCAACAUGUCGUUGGUGGUCUCCAGCAGGACCAUCACGUCCUGCUU ....(((.((((((......(((((..(((((((((((((.....))).....((.((.....)).)))))).)))))).)))))..((((((((......)))))))))))))).))). ( -46.00) >consensus GCUCAGCUGGAUGUAGUAGUCGAUGCCGUUGCGCAACUGCUCCUCGCAUUUCUGCAGCUUCUCGUCGCGCAUGCGCAGCAUAUCAUUGGUGGUCUCCAGCAGCACCAUCACGUCCUGCUU ....(((.((((((.......((((..((((((((..(((.....(((....))).((........))))))))))))).))))...((((((((.....)).)))))))))))).))). (-38.40 = -38.72 + 0.32)

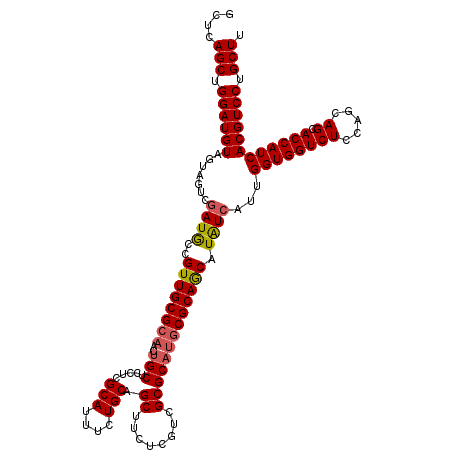

| Location | 15,759,002 – 15,759,122 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.42 |
| Mean single sequence MFE | -37.66 |
| Consensus MFE | -35.14 |
| Energy contribution | -35.38 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15759002 120 + 27905053 UAUCAUUGGUGGUCUCCAGCAGCACCAUCACGUCCUGCUUCUCCAGCUCGAUCUUGAUCAGCUCGUUCUCGGCCAGCUUCUGGCGCUUGUCCAGCUCGGCGCGCCAGUAGCUCUUCUCGC .........(((((...(((((.((......)).))))).....((((.(((....))))))).......)))))(((.(((((((..(((......)))))))))).)))......... ( -37.10) >DroSec_CAF1 16198 120 + 1 UAUCAUUGGUGGUCUCCAGCAGCACCAUCACGUCCUGCUUCUCCAGCUCGAUCUUGAUCAGCUCGUUCUCGGCCAGCUUCUGGCGCUUGUCCAGCUCGGCGCGCCAGUAGCUCUUCUCGC .........(((((...(((((.((......)).))))).....((((.(((....))))))).......)))))(((.(((((((..(((......)))))))))).)))......... ( -37.10) >DroSim_CAF1 16156 120 + 1 UAUCAUUGGUGGUCUCCAGCAGCACCAUCACGUCCUGCUUCUCCAGCUCGAUCUUGAUCAGCUCGUUCUCGGCCAGCUUCUGGCGCUUGUCCAGCUCGGCGCGCCAGUAGCUCUUCUCGC .........(((((...(((((.((......)).))))).....((((.(((....))))))).......)))))(((.(((((((..(((......)))))))))).)))......... ( -37.10) >DroEre_CAF1 18598 120 + 1 UAUCAUUGGUGGUCUCCAGCAGCACCAUCACGUCCUGCUUCUCCAGCUCGAUCUUGAUCAGCUCGUUCUCGGCCAGCUUCUGGCGCUUGUCCAGCUCGGCGCGCCAGUAGCUCUUCUCAC .........(((((...(((((.((......)).))))).....((((.(((....))))))).......)))))(((.(((((((..(((......)))))))))).)))......... ( -37.10) >DroPer_CAF1 16340 120 + 1 UGUCGUUGGUGGUCUCCAGCAGGACCAUCACGUCCUGCUUCUCCAGCUCGAUCUUGAUCAGCUCGUUCUCGGCUAGCUUCUGGCGCUUGUCCAGCUCCGCCCGCCAGUAGCUCUUCUCCC .((..(((((((.....((((((((......))))))))......((..((.((.(((.(((.((....))(((((...)))))))).))).)).)).)))))))))..))......... ( -39.90) >consensus UAUCAUUGGUGGUCUCCAGCAGCACCAUCACGUCCUGCUUCUCCAGCUCGAUCUUGAUCAGCUCGUUCUCGGCCAGCUUCUGGCGCUUGUCCAGCUCGGCGCGCCAGUAGCUCUUCUCGC .........(((((...(((((.((......)).))))).....((((.(((....))))))).......)))))(((.(((((((..(((......)))))))))).)))......... (-35.14 = -35.38 + 0.24)

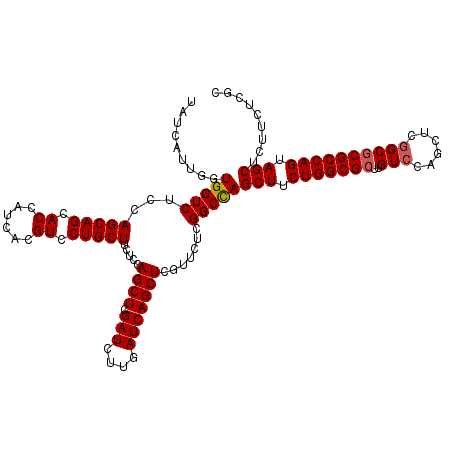

| Location | 15,759,002 – 15,759,122 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.42 |
| Mean single sequence MFE | -41.66 |
| Consensus MFE | -37.84 |
| Energy contribution | -38.24 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.91 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.516783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15759002 120 - 27905053 GCGAGAAGAGCUACUGGCGCGCCGAGCUGGACAAGCGCCAGAAGCUGGCCGAGAACGAGCUGAUCAAGAUCGAGCUGGAGAAGCAGGACGUGAUGGUGCUGCUGGAGACCACCAAUGAUA ((..((..((((.(((((((.((.....))....))))))).))))(((((....)).)))........))..))(((...(((((.((......)).)))))(....)..)))...... ( -41.50) >DroSec_CAF1 16198 120 - 1 GCGAGAAGAGCUACUGGCGCGCCGAGCUGGACAAGCGCCAGAAGCUGGCCGAGAACGAGCUGAUCAAGAUCGAGCUGGAGAAGCAGGACGUGAUGGUGCUGCUGGAGACCACCAAUGAUA ((..((..((((.(((((((.((.....))....))))))).))))(((((....)).)))........))..))(((...(((((.((......)).)))))(....)..)))...... ( -41.50) >DroSim_CAF1 16156 120 - 1 GCGAGAAGAGCUACUGGCGCGCCGAGCUGGACAAGCGCCAGAAGCUGGCCGAGAACGAGCUGAUCAAGAUCGAGCUGGAGAAGCAGGACGUGAUGGUGCUGCUGGAGACCACCAAUGAUA ((..((..((((.(((((((.((.....))....))))))).))))(((((....)).)))........))..))(((...(((((.((......)).)))))(....)..)))...... ( -41.50) >DroEre_CAF1 18598 120 - 1 GUGAGAAGAGCUACUGGCGCGCCGAGCUGGACAAGCGCCAGAAGCUGGCCGAGAACGAGCUGAUCAAGAUCGAGCUGGAGAAGCAGGACGUGAUGGUGCUGCUGGAGACCACCAAUGAUA (((.....((((.(((((((.((.....))....))))))).))))..((.....(.(((((((....))).)))).)...(((((.((......)).)))))))....)))........ ( -42.30) >DroPer_CAF1 16340 120 - 1 GGGAGAAGAGCUACUGGCGGGCGGAGCUGGACAAGCGCCAGAAGCUAGCCGAGAACGAGCUGAUCAAGAUCGAGCUGGAGAAGCAGGACGUGAUGGUCCUGCUGGAGACCACCAACGACA ((..(...((((.((((((...(........)...)))))).))))..((.....(.(((((((....))).)))).)...((((((((......))))))))))...)..))....... ( -41.50) >consensus GCGAGAAGAGCUACUGGCGCGCCGAGCUGGACAAGCGCCAGAAGCUGGCCGAGAACGAGCUGAUCAAGAUCGAGCUGGAGAAGCAGGACGUGAUGGUGCUGCUGGAGACCACCAAUGAUA ........((((.(((((((.((.....))....))))))).))))..((.....(.(((((((....))).)))).)...(((((.((......)).)))))))............... (-37.84 = -38.24 + 0.40)



| Location | 15,759,042 – 15,759,162 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -42.00 |
| Consensus MFE | -40.97 |
| Energy contribution | -41.23 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.822286 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15759042 120 + 27905053 CUCCAGCUCGAUCUUGAUCAGCUCGUUCUCGGCCAGCUUCUGGCGCUUGUCCAGCUCGGCGCGCCAGUAGCUCUUCUCGCGCUCCACCGCGCUCUCUAGGUCGCUCUCAACCUGCUGCUG ...((((..(((....)))(((..(((..(((((((((.(((((((..(((......)))))))))).))))......((((......))))......))))).....)))..))))))) ( -43.20) >DroSec_CAF1 16238 120 + 1 CUCCAGCUCGAUCUUGAUCAGCUCGUUCUCGGCCAGCUUCUGGCGCUUGUCCAGCUCGGCGCGCCAGUAGCUCUUCUCGCGCUCCACGGCGCUCUCCAGGUCGCUUUCCACCUGCUGCUG ...((((..(((....)))(((..((...(((((((((.(((((((..(((......)))))))))).))))......(((((....)))))......)))))......))..))))))) ( -42.70) >DroSim_CAF1 16196 120 + 1 CUCCAGCUCGAUCUUGAUCAGCUCGUUCUCGGCCAGCUUCUGGCGCUUGUCCAGCUCGGCGCGCCAGUAGCUCUUCUCGCGCUCCACGGCGCUCUCCAGGUCGCUCUCCACCUGCUGCUG ...((((..(((....)))(((..((...(((((((((.(((((((..(((......)))))))))).))))......(((((....)))))......)))))......))..))))))) ( -42.70) >DroEre_CAF1 18638 120 + 1 CUCCAGCUCGAUCUUGAUCAGCUCGUUCUCGGCCAGCUUCUGGCGCUUGUCCAGCUCGGCGCGCCAGUAGCUCUUCUCACGCUCCACGGCGCUCUCCAGGUCGUUCUCCACCUGCUGCUG ...((((..(((....)))(((..((...(((((((((.(((((((..(((......)))))))))).)))).......((((....)))).......)))))......))..))))))) ( -39.40) >consensus CUCCAGCUCGAUCUUGAUCAGCUCGUUCUCGGCCAGCUUCUGGCGCUUGUCCAGCUCGGCGCGCCAGUAGCUCUUCUCGCGCUCCACGGCGCUCUCCAGGUCGCUCUCCACCUGCUGCUG ...((((..(((....)))(((..((...(((((((((.(((((((..(((......)))))))))).))))......((((......))))......)))))......))..))))))) (-40.97 = -41.23 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:16 2006