
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,739,291 – 15,739,412 |
| Length | 121 |
| Max. P | 0.800089 |

| Location | 15,739,291 – 15,739,382 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 75.64 |
| Mean single sequence MFE | -16.05 |
| Consensus MFE | -7.47 |
| Energy contribution | -7.37 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.47 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.508446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
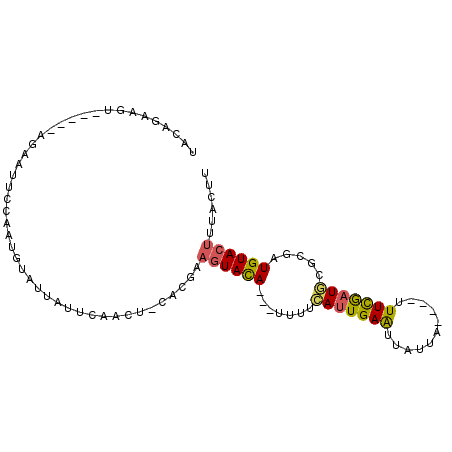
>3R_DroMel_CAF1 15739291 91 + 27905053 UACAGAAAUUACAGAAAAUUCCAAUGUAUUAUUCAACU-CACGAAGUACA---UUUUCAUUGAAUUAUCA----UUUCGCUGCGCGAUGUACUUUACUU ....(((..((((...........))))...)))....-...((((((((---((..((.((((......----.)))).))...)))))))))).... ( -14.60) >DroVir_CAF1 10816 91 + 1 -AAAGAAGUAAAGCAAAACUCGAGCGUAUUUUCUAUGUACAUAUAGUAUAUAGUUUUAAUUGAGGUCUUAUCAAUUUUAAUU--UGAUGUA-----CUC -.....((((........(((((...((....((((((((.....))))))))...)).)))))....((((((.......)--)))))))-----)). ( -17.10) >DroSec_CAF1 7852 86 + 1 UACAGAUGU-----AGAAUUCCAAUGUAUUAUUCAACU-CACUAAGUACA---UUUUCAUUGAAUUAACA----UUUCGAUGCGCGAUGUACUUUACCU ....((.((-----.((((...........)))).)))-)...(((((((---((..(((((((......----.)))))))...)))))))))..... ( -16.70) >DroSim_CAF1 7939 86 + 1 UACAGAAGU-----AGAAUUCCAAUGUAUUAUUCAACU-CACUAAGUACA---UUUUCAUUGAAUUAUAA----UUUCGAUGCGCGAUGUACUUUACCU ....((.((-----.((((...........)))).)))-)...(((((((---((..(((((((......----.)))))))...)))))))))..... ( -16.70) >DroEre_CAF1 7823 86 + 1 UACAGAAGU-----AGAAUUCCAAUGUAUUAUUCAACU-CACCAAGUACA---UUUUGAUUGAAUUAUUA----UUUCGAUGUGCGAUGUACUACUCUU ....((.((-----.((((...........)))).)))-)....((((((---((...((((((......----.))))))....))))))))...... ( -13.30) >DroYak_CAF1 8001 86 + 1 UACAGCAGU-----AGAAUUCCAAUGUAUUAUUCAACU-CAUGAAGUACA---UUAUCAUUGAAUUAUUA----UUUCGAUGUACGAUGUACUAUACUU ..((..(((-----.((((...........)))).)))-..)).((((((---((..(((((((......----.)))))))...))))))))...... ( -17.90) >consensus UACAGAAGU_____AGAAUUCCAAUGUAUUAUUCAACU_CACGAAGUACA___UUUUCAUUGAAUUAUUA____UUUCGAUGCGCGAUGUACUUUACUU ............................................((((((.......(((((((...........))))))).....))))))...... ( -7.47 = -7.37 + -0.11)

| Location | 15,739,316 – 15,739,412 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 83.26 |
| Mean single sequence MFE | -17.66 |
| Consensus MFE | -13.69 |
| Energy contribution | -14.42 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.800089 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15739316 96 + 27905053 GUAUUAUUCAACUCACGAAGUACAUUUUCAUUGAAUUAUCAUUUCGCUGCGCGAUGUACUUUACUUUGUAUCUUUGCUUUAUUAAUUUGUAAUGCA (((((((.........((((((((((..((.((((.......)))).))...)))))))))).....(((....)))...........))))))). ( -17.60) >DroSec_CAF1 7872 96 + 1 GUAUUAUUCAACUCACUAAGUACAUUUUCAUUGAAUUAACAUUUCGAUGCGCGAUGUACUUUACCUUGUAUCUUUGCUUUAUUAAUUUGUAAUGCA (((((((..........(((((((((..(((((((.......)))))))...)))))))))......(((....)))...........))))))). ( -19.70) >DroSim_CAF1 7959 96 + 1 GUAUUAUUCAACUCACUAAGUACAUUUUCAUUGAAUUAUAAUUUCGAUGCGCGAUGUACUUUACCUUGUAUCUUUGCUUUAUUAAUUUGUAAUGCA (((((((..........(((((((((..(((((((.......)))))))...)))))))))......(((....)))...........))))))). ( -19.70) >DroEre_CAF1 7843 96 + 1 GUAUUAUUCAACUCACCAAGUACAUUUUGAUUGAAUUAUUAUUUCGAUGUGCGAUGUACUACUCUUUGUAUCUUUGCUUUAUUAAUUUGUAAUGCA (((((((...........((((((((...((((((.......))))))....)))))))).......(((....)))...........))))))). ( -16.30) >DroYak_CAF1 8021 96 + 1 GUAUUAUUCAACUCAUGAAGUACAUUAUCAUUGAAUUAUUAUUUCGAUGUACGAUGUACUAUACUUUGUAUCUUUGCUUUAUUAAUUUGUAAUGCA (((((((.......((((((((((((..(((((((.......)))))))...)))))))).......(((....))).))))......))))))). ( -20.32) >DroAna_CAF1 8008 75 + 1 G-----------------AUUACAUCUUCUGUGAGUUA---CUUGGAUGCAUGAUGUAUUAUGCUUUGUAU-UUUGCUUUAUUAAUAUGUAAUGCA .-----------------(((((((.......((((..---..((.(.((((((....))))))..).)).-...)))).......)))))))... ( -12.34) >consensus GUAUUAUUCAACUCACGAAGUACAUUUUCAUUGAAUUAUCAUUUCGAUGCGCGAUGUACUAUACUUUGUAUCUUUGCUUUAUUAAUUUGUAAUGCA ..................((((((((..(((((((.......)))))))...))))))))..............(((.((((......)))).))) (-13.69 = -14.42 + 0.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:09:03 2006