| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,699,546 – 15,699,636 |
| Length | 90 |
| Max. P | 0.838245 |

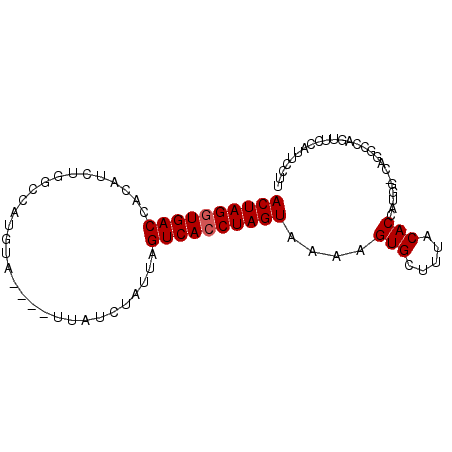
| Location | 15,699,546 – 15,699,636 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 83.28 |
| Mean single sequence MFE | -21.58 |
| Consensus MFE | -16.29 |
| Energy contribution | -16.69 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15699546 90 + 27905053 AAGGAAUGGAAUUGGCCUGGCCAUUGUGUAAAGCACUUUUACUAGCUGACUAAUAGAUAAUAAAUACAUGGUCAGAUGUGGUCACCUAGU .(((...((......))(((((((.(((.....))).........(((((((................)))))))..))))))))))... ( -20.09) >DroSec_CAF1 9013 86 + 1 AAGGAAUGGAACUGGCCUGUCCAUGGUGUAAAGCACUUUUACUAGGUGACUAAUAGAUAA----UACAUGGCCAGAUGUGGUCACCUAGU ..(((..((......))..)))..((((.....))))...(((((((((((.........----.((((......))))))))))))))) ( -24.30) >DroSim_CAF1 9048 86 + 1 AAGGAAUGGAACUGGCCUGUCCAUGGUGUAAAGCACUUUUACUAGGUGACUAAUAGAUAA----UACAUGGCCAGAUGUGGUCACCUAGU ..(((..((......))..)))..((((.....))))...(((((((((((.........----.((((......))))))))))))))) ( -24.30) >DroEre_CAF1 9234 71 + 1 UGGGAAUGGAACUGG-----GCAUGGUGUAAAGCACUUUUACUAGGUGACUAAUAGAUG-----------GC---AUGUGGUCACCUAGU ...............-----....((((.....))))...((((((((((((...(...-----------.)---...)))))))))))) ( -19.60) >DroYak_CAF1 9214 71 + 1 AAGGAAUGGAACUGG-----GCAUGGUGUAAAGCACUUUUACUAGGUGACUAAUAGAUG-----------GC---AUGUGGUCACCUAGU ...............-----....((((.....))))...((((((((((((...(...-----------.)---...)))))))))))) ( -19.60) >consensus AAGGAAUGGAACUGGCCUG_CCAUGGUGUAAAGCACUUUUACUAGGUGACUAAUAGAUAA____UACAUGGCCAGAUGUGGUCACCUAGU ........................((((.....))))...((((((((((((..........................)))))))))))) (-16.29 = -16.69 + 0.40)

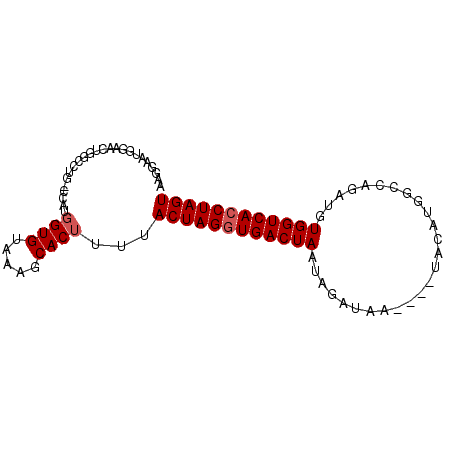
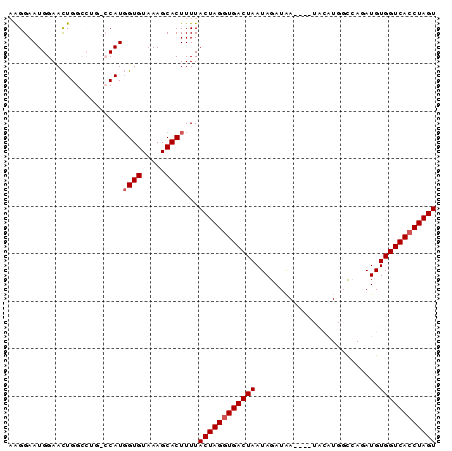
| Location | 15,699,546 – 15,699,636 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 83.28 |
| Mean single sequence MFE | -16.53 |
| Consensus MFE | -13.09 |
| Energy contribution | -13.29 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15699546 90 - 27905053 ACUAGGUGACCACAUCUGACCAUGUAUUUAUUAUCUAUUAGUCAGCUAGUAAAAGUGCUUUACACAAUGGCCAGGCCAAUUCCAUUCCUU (((((.((((.......((..(((....)))..)).....)))).)))))....(((.....)))..((((...))))............ ( -15.50) >DroSec_CAF1 9013 86 - 1 ACUAGGUGACCACAUCUGGCCAUGUA----UUAUCUAUUAGUCACCUAGUAAAAGUGCUUUACACCAUGGACAGGCCAGUUCCAUUCCUU ...(((.........((((((.(((.----.....((((((....))))))...(((.....))).....))))))))).......))). ( -18.69) >DroSim_CAF1 9048 86 - 1 ACUAGGUGACCACAUCUGGCCAUGUA----UUAUCUAUUAGUCACCUAGUAAAAGUGCUUUACACCAUGGACAGGCCAGUUCCAUUCCUU ...(((.........((((((.(((.----.....((((((....))))))...(((.....))).....))))))))).......))). ( -18.69) >DroEre_CAF1 9234 71 - 1 ACUAGGUGACCACAU---GC-----------CAUCUAUUAGUCACCUAGUAAAAGUGCUUUACACCAUGC-----CCAGUUCCAUUCCCA ((((((((((.....---..-----------.........))))))))))....(((.....))).....-----............... ( -14.89) >DroYak_CAF1 9214 71 - 1 ACUAGGUGACCACAU---GC-----------CAUCUAUUAGUCACCUAGUAAAAGUGCUUUACACCAUGC-----CCAGUUCCAUUCCUU ((((((((((.....---..-----------.........))))))))))....(((.....))).....-----............... ( -14.89) >consensus ACUAGGUGACCACAUCUGGCCAUGUA____UUAUCUAUUAGUCACCUAGUAAAAGUGCUUUACACCAUGG_CAGGCCAGUUCCAUUCCUU ((((((((((..............................))))))))))....(((.....)))......................... (-13.09 = -13.29 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:45 2006