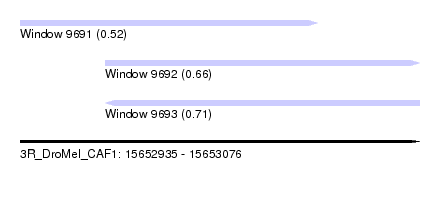
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,652,935 – 15,653,076 |
| Length | 141 |
| Max. P | 0.714542 |

| Location | 15,652,935 – 15,653,040 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 88.10 |
| Mean single sequence MFE | -24.48 |
| Consensus MFE | -20.31 |
| Energy contribution | -20.88 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15652935 105 + 27905053 UUUCAUAUGGCCAUUGGCAUUAGUUUAUUUGGUUGUUCAUGCAGUCCAAGUGGAAUGGGUGGUUCGAAGUUCGAAACAAAAAAACCAAACAAA-GAACCCCCAAAG .........(((...))).....(((((((((((((....)))).))))))))).((((.(((((...(((.(............).)))...-)))))))))... ( -26.90) >DroSim_CAF1 175855 105 + 1 UUUCAUAUGGCCAUUGGCAUUAGUUUAUUUGGUUGUUCAUGCAGUCCAAGUGGAAUGGGUGGUUCGAACUUCGAAACAAA-AAACCAAACAAAGGAACCCCCAAAG .........(((...))).....(((((((((((((....)))).))))))))).((((.((((((.....)))......-...((.......)).)))))))... ( -25.30) >DroEre_CAF1 158728 91 + 1 UUUCAUAUGGCCAUUGGCAUUAGUUUAUUUGGCUCUUCAUGCAGUCCAA-UGGAAUGGGUGGUUCGAA----------AA-AAACCAAACAA---AACCCCCGAAG .(((......((((((((..(((.....)))((.......)).).))))-)))...(((.((((....----------..-...........---)))))))))). ( -18.33) >DroYak_CAF1 183023 95 + 1 UUUCAUAUGGCCAUUGGCAUUAGUUUAUUUGGUUGUUCAUGCAGUCCAAGUGGAAUGGGUGGUUCGAA----------AA-AAACCAAACCAAAGAACCACCGAAG .(((.....(((...))).....(((((((((((((....)))).)))))))))...((((((((...----------..-.............))))))))))). ( -27.37) >consensus UUUCAUAUGGCCAUUGGCAUUAGUUUAUUUGGUUGUUCAUGCAGUCCAAGUGGAAUGGGUGGUUCGAA__________AA_AAACCAAACAAA_GAACCCCCAAAG .........(((...))).....(((((((((((((....)))).))))))))).((((.(((((.............................)))))))))... (-20.31 = -20.88 + 0.56)



| Location | 15,652,965 – 15,653,076 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 87.13 |
| Mean single sequence MFE | -19.10 |
| Consensus MFE | -15.90 |
| Energy contribution | -16.35 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.655109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15652965 111 + 27905053 GGUUGUUCAUGCAGUCCAAGUGGAAUGGGUGGUUCGAAGUUCGAAACAAAAAAACCAAACAAA-GAACCCCCAAAGAAAUUGAACAUAAAACAUAUUCAAACCGAAAUACAA (((((((((.....(((....))).((((.(((((...(((.(............).)))...-))))))))).......))))))..............)))......... ( -22.23) >DroSim_CAF1 175885 111 + 1 GGUUGUUCAUGCAGUCCAAGUGGAAUGGGUGGUUCGAACUUCGAAACAAA-AAACCAAACAAAGGAACCCCCAAAGAAAUUGAACAUAAAACAUAUUCAAACCGAAAUAAAA (((((((((.....(((....))).((((.((((((.....)))......-...((.......)).))))))).......))))))..............)))......... ( -20.63) >DroEre_CAF1 158758 97 + 1 GGCUCUUCAUGCAGUCCAA-UGGAAUGGGUGGUUCGAA----------AA-AAACCAAACAA---AACCCCCGAAGAAACUGAACAUUAAACAUAUUCAAACCGAAAUACAA ((.(((((......(((..-.)))..(((.((((....----------..-...........---))))))))))))...((((.((.....)).))))..))......... ( -14.43) >consensus GGUUGUUCAUGCAGUCCAAGUGGAAUGGGUGGUUCGAA_UUCGAAACAAA_AAACCAAACAAA_GAACCCCCAAAGAAAUUGAACAUAAAACAUAUUCAAACCGAAAUACAA ...((((((.....(((....))).((((.(((((.............................))))))))).......)))))).......................... (-15.90 = -16.35 + 0.45)


| Location | 15,652,965 – 15,653,076 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 87.13 |
| Mean single sequence MFE | -25.79 |
| Consensus MFE | -18.59 |
| Energy contribution | -18.82 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15652965 111 - 27905053 UUGUAUUUCGGUUUGAAUAUGUUUUAUGUUCAAUUUCUUUGGGGGUUC-UUUGUUUGGUUUUUUUGUUUCGAACUUCGAACCACCCAUUCCACUUGGACUGCAUGAACAACC .........((((.(..(((((.....((((((......(((((((((-...((((((..........))))))...))))).))))......)))))).)))))..))))) ( -28.50) >DroSim_CAF1 175885 111 - 1 UUUUAUUUCGGUUUGAAUAUGUUUUAUGUUCAAUUUCUUUGGGGGUUCCUUUGUUUGGUUU-UUUGUUUCGAAGUUCGAACCACCCAUUCCACUUGGACUGCAUGAACAACC .........((((.(..(((((.....((((((......((((((((((((((........-.......)))))...))))).))))......)))))).)))))..))))) ( -24.96) >DroEre_CAF1 158758 97 - 1 UUGUAUUUCGGUUUGAAUAUGUUUAAUGUUCAGUUUCUUCGGGGGUU---UUGUUUGGUUU-UU----------UUCGAACCACCCAUUCCA-UUGGACUGCAUGAAGAGCC .........(((((...(((((.....(((((((..(....)((((.---..((((((...-..----------.)))))).)))).....)-)))))).)))))..))))) ( -23.90) >consensus UUGUAUUUCGGUUUGAAUAUGUUUUAUGUUCAAUUUCUUUGGGGGUUC_UUUGUUUGGUUU_UUUGUUUCGAA_UUCGAACCACCCAUUCCACUUGGACUGCAUGAACAACC .........((((....(((((.....((((((......(((((((((.............................))))).))))......)))))).)))))...)))) (-18.59 = -18.82 + 0.23)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:28 2006