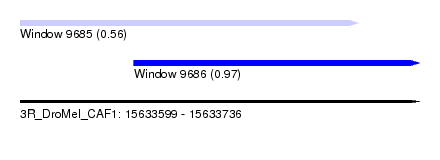
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,633,599 – 15,633,736 |
| Length | 137 |
| Max. P | 0.968473 |

| Location | 15,633,599 – 15,633,715 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 82.70 |
| Mean single sequence MFE | -30.78 |
| Consensus MFE | -19.31 |
| Energy contribution | -18.89 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.556031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15633599 116 + 27905053 UAAAUGCAUUUGCAGCUGUGUUUGU-GCGGGGGGGCGUAAAUAGUACAGAGCGUGCUUAGUUGCGUUCGCAAUUAAUUCACAGUAAACAGCAAAUGUUGCAAAUAAAUUAGCAGCGA ....(((....)))(((((..((((-(...(.((((((((..(((((.....)))))...)))))))).)........)))))...)))))...((((((..........)))))). ( -35.50) >DroVir_CAF1 286963 101 + 1 ----UGUAUCUGCAGCUGUGUUUGU-GCG----CGCGUAAAUAGUGCAAAGCGCGCUUAGCUGCAUUUGCAAUUAAUUCACAG-------CAAAUGUUGCAAAUAAAUUAGCAGGCC ----....((((((((.((((((.(-(((----(.........))))))))))))))..((.((((((((............)-------))))))).))..........))))).. ( -32.90) >DroPse_CAF1 172551 116 + 1 UAAAUGUAUCUGUAUCUGUAUCUGUUGUGUG-GGGCGUAAAUAGUGCAGAGUGCGCUUAGUGGCAUUUGCAAUUAAUUCACAGUAAACAGCAAAUGUUGCAAAUAAAUUAGCAGCAA .........((((..((((........((..-((((.((...(((((.....)))))...)))).))..))........))))...))))....((((((..........)))))). ( -25.49) >DroMoj_CAF1 262321 108 + 1 ----CAUAGCUGCAGCUGUGUUUGU-GCG----CGCGUAAAUAGCGCAAAGCGCGCUUAGUGGCAUUUGCAAUUAAUUCACAGCAAACAGCAAAUGUUGCAAAUAAAUUAGCAGGCC ----.....((((.((((((...((-(((----(((((.....))))...))))))(((((.((....)).)))))..))))))...((((....))))...........))))... ( -35.80) >DroAna_CAF1 154904 113 + 1 UAAAUGUAUCUGCAUCUGUGUUUGU-GCGG---GGCGUAAAUAGUGCAGAGCGUGCUUAGUUGCGUUUGCAAUUAAUUCACAGUAAACAGCAAAUGUUGCAAAUAAAUUAGCAGCAA ....(((.(((((((...(((((((-((..---.))))))))))))))))..(((.((((((((....))))))))..)))........)))..((((((..........)))))). ( -29.70) >DroPer_CAF1 190878 110 + 1 UAAAUGUAUCU------GUAUCUGUUGUGUG-GGGCGUAAAUAGUGCAGAGUGCGCUUAGUGGCAUUUGCAAUUAAUUCACAGUAAACAGCAAAUGUUGCAAAUAAAUUAGCAGCAA ...........------.....((((((...-(((((((............))))))).(..((((((((...................))))))))..)..........)))))). ( -25.31) >consensus UAAAUGUAUCUGCAGCUGUGUUUGU_GCG____GGCGUAAAUAGUGCAGAGCGCGCUUAGUGGCAUUUGCAAUUAAUUCACAGUAAACAGCAAAUGUUGCAAAUAAAUUAGCAGCAA .........((((.....(((((((.((......))......(((((.....))))).....((((((((...................)))))))).))))))).....))))... (-19.31 = -18.89 + -0.41)



| Location | 15,633,638 – 15,633,736 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 79.66 |
| Mean single sequence MFE | -22.36 |
| Consensus MFE | -16.41 |
| Energy contribution | -15.66 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15633638 98 + 27905053 AUAGUACAGAGCGUGCUUAGUUGCGUUCGCAAUUAAUUCACAGUAAACAGCAAAUGUUGCAAAUAAAUUAGCAGCGACAAA---AAGUGUCUGCAAGUGAA ...((((.....))))((((((((....))))))))(((((.(....).(((...(((((..........)))))((((..---...)))))))..))))) ( -24.50) >DroPse_CAF1 172590 84 + 1 AUAGUGCAGAGUGCGCUUAGUGGCAUUUGCAAUUAAUUCACAGUAAACAGCAAAUGUUGCAAAUAAAUUAGCAGCAACAAA---AAC-------------- ..(((((.....)))))..(..((((((((...................))))))))..).....................---...-------------- ( -18.51) >DroGri_CAF1 189848 90 + 1 AUAGCGCAAAUCGCGCUUAGUUGCAUUUGCAAUUAAUGCAAUGCAAACAGCGAAUGUUGCAAAUAAAUUAGCAGGCUAAAA---AAC-------A-ACAAU .....((((....((((...(((((((.(((.....))))))))))..))))....)))).......((((....))))..---...-------.-..... ( -23.60) >DroMoj_CAF1 262352 92 + 1 AUAGCGCAAAGCGCGCUUAGUGGCAUUUGCAAUUAAUUCACAGCAAACAGCAAAUGUUGCAAAUAAAUUAGCAGGCCAAAU-GCAGC-------A-ACAAC ...((((...))))(((..(..((((((((...................))))))))..)..........(((.......)-)))))-------.-..... ( -23.91) >DroAna_CAF1 154940 99 + 1 AUAGUGCAGAGCGUGCUUAGUUGCGUUUGCAAUUAAUUCACAGUAAACAGCAAAUGUUGCAAAUAAAUUAGCAGCAACAAAA--AAGUAUCUGCAAGUGAA ....(((((((((((.((((((((....))))))))..))).(....).))...((((((..........))))))......--.....))))))...... ( -25.10) >DroPer_CAF1 190911 84 + 1 AUAGUGCAGAGUGCGCUUAGUGGCAUUUGCAAUUAAUUCACAGUAAACAGCAAAUGUUGCAAAUAAAUUAGCAGCAACAAA---AAC-------------- ..(((((.....)))))..(..((((((((...................))))))))..).....................---...-------------- ( -18.51) >consensus AUAGUGCAGAGCGCGCUUAGUGGCAUUUGCAAUUAAUUCACAGUAAACAGCAAAUGUUGCAAAUAAAUUAGCAGCAACAAA___AAC_______A____A_ ..(((((.....)))))..(..((((((((...................))))))))..)......................................... (-16.41 = -15.66 + -0.75)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:08:22 2006