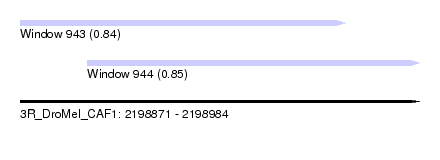
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,198,871 – 2,198,984 |
| Length | 113 |
| Max. P | 0.852322 |

| Location | 2,198,871 – 2,198,963 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 81.30 |
| Mean single sequence MFE | -38.48 |
| Consensus MFE | -25.83 |
| Energy contribution | -26.50 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
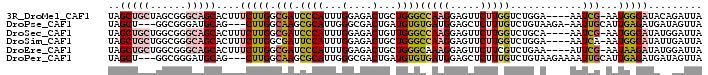
>3R_DroMel_CAF1 2198871 92 + 27905053 ----GGU-----------CUUCUGGACGGUGCUGUAGCUGCUAGCGGGCAGCACUUUCUUGGCGAUCCCAUUUGGAGACUGCUGGGCCAAGGAGUUCUUGGUCUGGA- ----(((-----------((((.(((.((((((((.((.....))..)))))))).)))(((.....)))...))))))).(..(((((((.....)))))))..).- ( -40.10) >DroSec_CAF1 1904 92 + 1 ----GGU-----------CUUCUGGACGGUGCUGUAGCUGCUGGCGGGCAGCACUUUCUUGGCGAUCCCAUUUGGAGACUGUUGGGCCAAGGAGUUCUUGGUCUGCA- ----(((-----------((((.(((.((((((((.((.....))..)))))))).)))(((.....)))...)))))))...((((((((.....))))))))...- ( -37.20) >DroSim_CAF1 1902 92 + 1 ----GGU-----------CUUCUGGACGGUGCUGUAGCUGCUGGCGGGCAGCACUUUCUUGGCGAUUCCAUUUGGAGACUGCUGGGCCAAGGAGUUCUUGGUCUGGA- ----(((-----------((((.(((.((((((((.((.....))..)))))))).)))(((.....)))...))))))).(..(((((((.....)))))))..).- ( -40.00) >DroEre_CAF1 2230 92 + 1 ----GGU-----------CUUCUGGGCGGUGCUGUAGCUGCUGGCGGGCAGCACUUUCUUGGCGAUCCCAUUUGGAGACUGCUGGGCAAAGGAGUUCUUCGUCUGAA- ----(((-----------(.((.(((.((((((((.((.....))..)))))))).))).)).)))).((..((((((((.((......)).)).))))))..))..- ( -32.60) >DroYak_CAF1 1906 92 + 1 ----GGU-----------CUUCUGGGCGGUGCUGUAGCUGCUGGCGGGCAGCACAUUCUUGGCGAUACCAUUUGGAGACUGCUGGGCCAAGGAGUUCUUGGUCUGGA- ----(((-----------((.(..((.((((.(((.((((((....)))))).((....))))).)))).))..)))))).(..(((((((.....)))))))..).- ( -37.80) >DroPer_CAF1 15599 102 + 1 UGAUGAUGGUGCGGGGGGUUCCUUCCUGCUGCUGUAGCU---GGCGGGAUGCAG---CUUGGCAAGCGCAUUGGGCGACUGAUGUGUGAUGGAGCUCUUUGUCUGUAA .....((((.((((((((((((.(((((((((....)).---)))))))(((..---....))).((((((..(....)..))))))...)))))))))))))))).. ( -43.20) >consensus ____GGU___________CUUCUGGACGGUGCUGUAGCUGCUGGCGGGCAGCACUUUCUUGGCGAUCCCAUUUGGAGACUGCUGGGCCAAGGAGUUCUUGGUCUGGA_ .......................((((.(((((((.((.....))..))))))).((((((((....(((...(....)...))))))))))).......)))).... (-25.83 = -26.50 + 0.67)

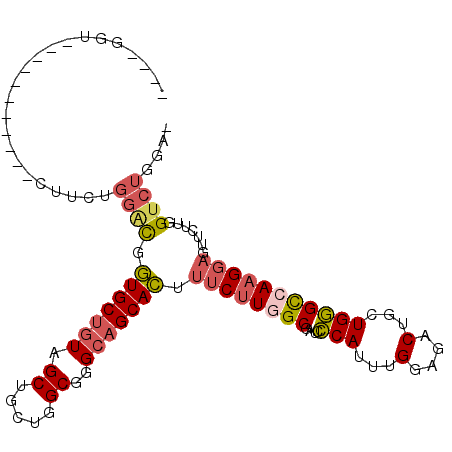
| Location | 2,198,890 – 2,198,984 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 77.65 |
| Mean single sequence MFE | -32.50 |
| Consensus MFE | -13.95 |
| Energy contribution | -13.98 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.852322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2198890 94 + 27905053 UAGCUGCUAGCGGGCAGCACUUUCUUGGCGAUCCCAUUUGGAGACUGCUGGGCCAAGGAGUUCUUGGUCUGGA----AAUCG-AAUGGGAUACAGAUUA ..((((((....))))))......(((...(((((((((((......(..(((((((.....)))))))..).----..)))-)))))))).))).... ( -39.00) >DroPse_CAF1 12235 92 + 1 UAGCU---GGCGGGAUGCAG---CUUGGCAAGCGCAUUGGGCGACUGAUGUGUGAUGGAGCUCUUUGUCUGUAAGA-AAUUGCAUUGAGAUGAUAGUUA (((((---(.(..(((((((---(((.(...((((((..(....)..))))))..).))))((((.......))))-...))))))..)....)))))) ( -28.80) >DroSec_CAF1 1923 94 + 1 UAGCUGCUGGCGGGCAGCACUUUCUUGGCGAUCCCAUUUGGAGACUGUUGGGCCAAGGAGUUCUUGGUCUGCA----AAUCG-AAUGGGAUAUGGAUUA ..((((((....))))))....(((.....(((((((((((....(((.((((((((.....)))))))))))----..)))-))))))))..)))... ( -35.80) >DroSim_CAF1 1921 94 + 1 UAGCUGCUGGCGGGCAGCACUUUCUUGGCGAUUCCAUUUGGAGACUGCUGGGCCAAGGAGUUCUUGGUCUGGA----AAUCA-AAUGGGAUAUUGAUUA ..((((((....))))))............(((((((((((......(..(((((((.....)))))))..).----..)))-))))))))........ ( -34.80) >DroEre_CAF1 2249 94 + 1 UAGCUGCUGGCGGGCAGCACUUUCUUGGCGAUCCCAUUUGGAGACUGCUGGGCAAAGGAGUUCUUCGUCUGAA----AUUCG-AAUAAGAUAUGGAUUA ..((((((....)))))).((((((..(((.((((....)).)).)))..)).)))).(((((...((((...----.....-....))))..))))). ( -27.80) >DroPer_CAF1 15633 93 + 1 UAGCU---GGCGGGAUGCAG---CUUGGCAAGCGCAUUGGGCGACUGAUGUGUGAUGGAGCUCUUUGUCUGUAAGAAAAUUGCAUUGAGAUGAUAGUUA (((((---(.(..(((((((---(((.(...((((((..(....)..))))))..).))))((((.......))))....))))))..)....)))))) ( -28.80) >consensus UAGCUGCUGGCGGGCAGCACUUUCUUGGCGAUCCCAUUUGGAGACUGCUGGGCCAAGGAGUUCUUGGUCUGGA____AAUCG_AAUGAGAUAUUGAUUA ..(((((......)))))....(((((.(((.((((...(....)...))))(((((.....)))))............)))...)))))......... (-13.95 = -13.98 + 0.03)
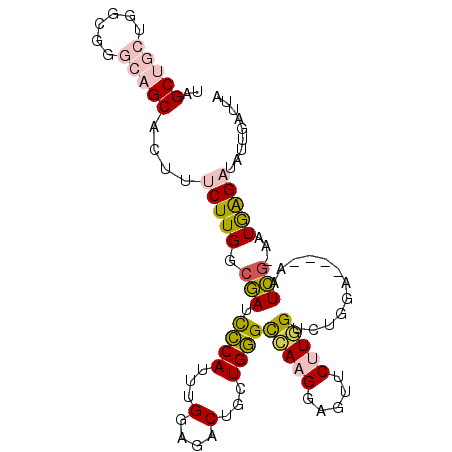
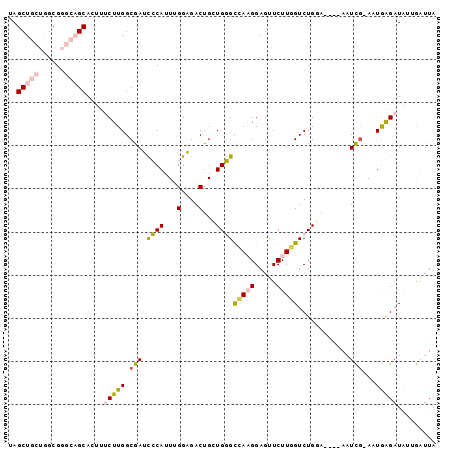
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:56 2006