
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,567,739 – 15,567,875 |
| Length | 136 |
| Max. P | 0.999404 |

| Location | 15,567,739 – 15,567,844 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 88.10 |
| Mean single sequence MFE | -36.89 |
| Consensus MFE | -34.98 |
| Energy contribution | -35.94 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.57 |
| SVM RNA-class probability | 0.999404 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15567739 105 + 27905053 CAUCCAAUCUACUCUUCAGUCUGUGAAAAUGCAAUCAAGUCGCUGA------AGCUGCAGCUGCAGCUGCUGUUGCAUCUGCAGCUUAAAUCGCCGGCGACACCAAAGGAG ..(((.............(..(((......)))..)..((((((((------((((((((.((((((....)))))).))))))))).......)))))))......))). ( -39.31) >DroSec_CAF1 92853 105 + 1 CAUCCAAUCUACUCUUCAGUCUGUGAAAAUGCAAUCAAGUCGCUGA------AGCUGCAGCUCCAGCUGCUGUUGCAUCUGCAGCUUAAAUCGCCGGCGACACCAAAGGAG ..(((.............(..(((......)))..)..((((((((------((((((((...((((....))))...))))))))).......)))))))......))). ( -32.91) >DroSim_CAF1 86875 105 + 1 CAUCCAAUCUACUCUUCAGUCUGUGAAAAUGCAAUCAAGUCGCUGA------AGCUGCAGCUGCAGCUGCUGUUGCAUCUGCAGCUUAAAUCGCCGGCGACACCAAAGGAG ..(((.............(..(((......)))..)..((((((((------((((((((.((((((....)))))).))))))))).......)))))))......))). ( -39.31) >DroEre_CAF1 82250 110 + 1 CAUCCAAUCUACUCUUCAGUCUGUGAAAAUGCAAUCAAGUCGCCGAGGCUGCAGCUGCAGCUGCAGCUGCUGUUGCAUCUGCAGCUCGAAUCGCCGGCGACACCA-AGGAG ..(((.............(..(((......)))..)..((((((..(((..(((((((((.((((((....)))))).)))))))).)....)))))))))....-.))). ( -43.60) >DroYak_CAF1 91447 105 + 1 UAUCCAAUCUUCUCUUCAGUCUGUGAAAAUGCAAUCAAGUCGCUGA------AGCUGCAGCUGCAGCUGCUGUUGCAUCUGCAGCUCAAAUCGCCGGCGACACCAAAGGAG ..(((.............(..(((......)))..)..(((((((.------((((((((.((((((....)))))).))))))))........)))))))......))). ( -37.40) >DroAna_CAF1 93739 92 + 1 CAUUCAAUCCGCUCUUCAGUCUGUGAAAAUGCAAUCAAGUCGCUGC------AGCUGCCACUGCGG------------GAGCAGCUUAAAUCGCCAGCGACACCAGUGGA- .......((((((.....(..(((......)))..)..((((((((------((((((..(....)------------..))))))......).)))))))...))))))- ( -28.80) >consensus CAUCCAAUCUACUCUUCAGUCUGUGAAAAUGCAAUCAAGUCGCUGA______AGCUGCAGCUGCAGCUGCUGUUGCAUCUGCAGCUUAAAUCGCCGGCGACACCAAAGGAG ..(((.............(..(((......)))..)..(((((((.......((((((((.((((((....)))))).))))))))........)))))))......))). (-34.98 = -35.94 + 0.96)



| Location | 15,567,739 – 15,567,844 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 88.10 |
| Mean single sequence MFE | -35.92 |
| Consensus MFE | -30.24 |
| Energy contribution | -30.99 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15567739 105 - 27905053 CUCCUUUGGUGUCGCCGGCGAUUUAAGCUGCAGAUGCAACAGCAGCUGCAGCUGCAGCU------UCAGCGACUUGAUUGCAUUUUCACAGACUGAAGAGUAGAUUGGAUG .(((.(..(.(((((.(((......((((((((.(((....))).))))))))...)))------...))))))..)((((.((((((.....))))))))))...))).. ( -36.60) >DroSec_CAF1 92853 105 - 1 CUCCUUUGGUGUCGCCGGCGAUUUAAGCUGCAGAUGCAACAGCAGCUGGAGCUGCAGCU------UCAGCGACUUGAUUGCAUUUUCACAGACUGAAGAGUAGAUUGGAUG ((.((((...(((((.(((.......)))))(((((((((((..((((((((....)))------)))))...))).)))))))).....)))...)))).))........ ( -34.50) >DroSim_CAF1 86875 105 - 1 CUCCUUUGGUGUCGCCGGCGAUUUAAGCUGCAGAUGCAACAGCAGCUGCAGCUGCAGCU------UCAGCGACUUGAUUGCAUUUUCACAGACUGAAGAGUAGAUUGGAUG .(((.(..(.(((((.(((......((((((((.(((....))).))))))))...)))------...))))))..)((((.((((((.....))))))))))...))).. ( -36.60) >DroEre_CAF1 82250 110 - 1 CUCCU-UGGUGUCGCCGGCGAUUCGAGCUGCAGAUGCAACAGCAGCUGCAGCUGCAGCUGCAGCCUCGGCGACUUGAUUGCAUUUUCACAGACUGAAGAGUAGAUUGGAUG .((((-..(.(((((((((......((((((((.(((....))).))))))))((....)).)))..)))))))..)((((.((((((.....))))))))))...))).. ( -43.20) >DroYak_CAF1 91447 105 - 1 CUCCUUUGGUGUCGCCGGCGAUUUGAGCUGCAGAUGCAACAGCAGCUGCAGCUGCAGCU------UCAGCGACUUGAUUGCAUUUUCACAGACUGAAGAGAAGAUUGGAUA .(((......(((((.(((....((((((((((.(((....))).)))))))).)))))------...)))))..((((...((((((.....))))))...))))))).. ( -35.20) >DroAna_CAF1 93739 92 - 1 -UCCACUGGUGUCGCUGGCGAUUUAAGCUGCUC------------CCGCAGUGGCAGCU------GCAGCGACUUGAUUGCAUUUUCACAGACUGAAGAGCGGAUUGAAUG -(((......(((((((........((((((..------------(....)..))))))------.)))))))......((.((((((.....)))))))))))....... ( -29.40) >consensus CUCCUUUGGUGUCGCCGGCGAUUUAAGCUGCAGAUGCAACAGCAGCUGCAGCUGCAGCU______UCAGCGACUUGAUUGCAUUUUCACAGACUGAAGAGUAGAUUGGAUG ..........(((((.((.......((((((((.((((........)))).))))))))......)).)))))(..(((...((((((.....))))))...)))..)... (-30.24 = -30.99 + 0.75)

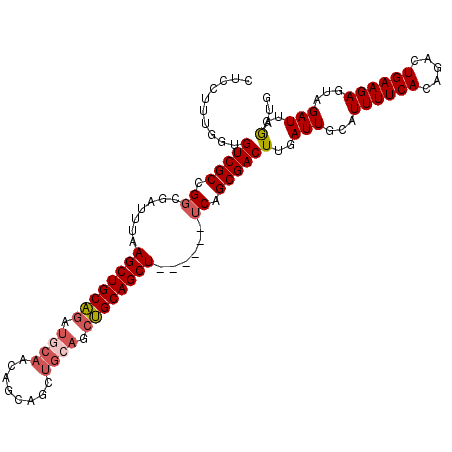

| Location | 15,567,775 – 15,567,875 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 85.82 |
| Mean single sequence MFE | -37.39 |
| Consensus MFE | -33.69 |
| Energy contribution | -34.79 |
| Covariance contribution | 1.10 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.46 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.995466 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15567775 100 + 27905053 AAGUCGCUGA------AGCUGCAGCUGCAGCUGCUGUUGCAUCUGCAGCUUAAAUCGCCGGCGACACCAAAGGAGAAGGCAGUGGCAACAAAAAGUGUCAAUUGAC ..((((((.(------((((((((.((((((....)))))).))))))))).....(((..(..(......)..)..)))))))))..........(((....))) ( -40.40) >DroSec_CAF1 92889 100 + 1 AAGUCGCUGA------AGCUGCAGCUCCAGCUGCUGUUGCAUCUGCAGCUUAAAUCGCCGGCGACACCAAAGGAGAAGGCAAUGGCAACAAAAAGUGUCAAUUGAC ..((((((((------((((((((...((((....))))...))))))))).......)))))))............((((.((....)).....))))....... ( -33.61) >DroSim_CAF1 86911 100 + 1 AAGUCGCUGA------AGCUGCAGCUGCAGCUGCUGUUGCAUCUGCAGCUUAAAUCGCCGGCGACACCAAAGGAGAAGGCAAUGGCAACAAAAAGUGUCAAUUGAC ..((((((((------((((((((.((((((....)))))).))))))))).......)))))))............((((.((....)).....))))....... ( -40.01) >DroEre_CAF1 82286 105 + 1 AAGUCGCCGAGGCUGCAGCUGCAGCUGCAGCUGCUGUUGCAUCUGCAGCUCGAAUCGCCGGCGACACCA-AGGAGAAGGCAACGGCAACAAAAAGUGUCAAUUGAC ..((((.(((((((((((.((((((.((....)).)))))).))))))))....))).))))(((((..-.......(....)(....).....)))))....... ( -44.00) >DroYak_CAF1 91483 100 + 1 AAGUCGCUGA------AGCUGCAGCUGCAGCUGCUGUUGCAUCUGCAGCUCAAAUCGCCGGCGACACCAAAGGAGAAGGCAACGGCAACAAAAAGUGUCAAUUGAC ..((((.(((------((((((((.((((((....)))))).))))))))....))).))))(((((..........(....)(....).....)))))....... ( -39.50) >DroAna_CAF1 93775 79 + 1 AAGUCGCUGC------AGCUGCCACUGCGG------------GAGCAGCUUAAAUCGCCAGCGACACCAGUGGA---------GGCAACAAAAAGUGUCAAUUGAC ..((((((((------((((((..(....)------------..))))))......).)))))))..((((.((---------.((........)).)).)))).. ( -26.80) >consensus AAGUCGCUGA______AGCUGCAGCUGCAGCUGCUGUUGCAUCUGCAGCUUAAAUCGCCGGCGACACCAAAGGAGAAGGCAAUGGCAACAAAAAGUGUCAAUUGAC ..(((((((.......((((((((.((((((....)))))).))))))))........)))))))..................((((........))))....... (-33.69 = -34.79 + 1.10)

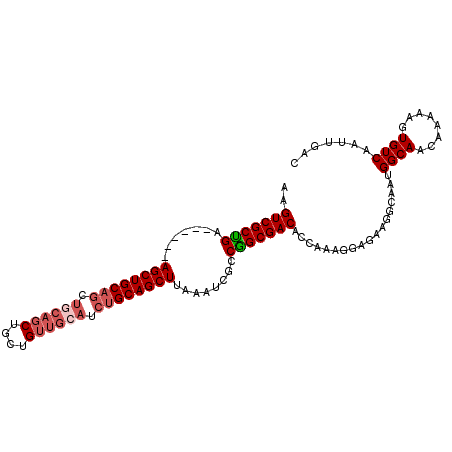

| Location | 15,567,775 – 15,567,875 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 85.82 |
| Mean single sequence MFE | -34.45 |
| Consensus MFE | -27.90 |
| Energy contribution | -28.79 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15567775 100 - 27905053 GUCAAUUGACACUUUUUGUUGCCACUGCCUUCUCCUUUGGUGUCGCCGGCGAUUUAAGCUGCAGAUGCAACAGCAGCUGCAGCUGCAGCU------UCAGCGACUU (((....)))..........((((..(.......)..))))(((((.(((......((((((((.(((....))).))))))))...)))------...))))).. ( -33.60) >DroSec_CAF1 92889 100 - 1 GUCAAUUGACACUUUUUGUUGCCAUUGCCUUCUCCUUUGGUGUCGCCGGCGAUUUAAGCUGCAGAUGCAACAGCAGCUGGAGCUGCAGCU------UCAGCGACUU (((............(((((((....(((.........)))(((..((((.......))))..))))))))))..((((((((....)))------)))))))).. ( -32.90) >DroSim_CAF1 86911 100 - 1 GUCAAUUGACACUUUUUGUUGCCAUUGCCUUCUCCUUUGGUGUCGCCGGCGAUUUAAGCUGCAGAUGCAACAGCAGCUGCAGCUGCAGCU------UCAGCGACUU (((....)))..........((((..(.......)..))))(((((.(((......((((((((.(((....))).))))))))...)))------...))))).. ( -34.40) >DroEre_CAF1 82286 105 - 1 GUCAAUUGACACUUUUUGUUGCCGUUGCCUUCUCCU-UGGUGUCGCCGGCGAUUCGAGCUGCAGAUGCAACAGCAGCUGCAGCUGCAGCUGCAGCCUCGGCGACUU (((....))).......(((((((((((..(((.((-(((.((((....)))))))))....))).))))).((((((((....))))))))......)))))).. ( -44.40) >DroYak_CAF1 91483 100 - 1 GUCAAUUGACACUUUUUGUUGCCGUUGCCUUCUCCUUUGGUGUCGCCGGCGAUUUGAGCUGCAGAUGCAACAGCAGCUGCAGCUGCAGCU------UCAGCGACUU (((....))).....((((((..(((((.............((((....))))...((((((((.(((....))).))))))))))))).------.))))))... ( -35.20) >DroAna_CAF1 93775 79 - 1 GUCAAUUGACACUUUUUGUUGCC---------UCCACUGGUGUCGCUGGCGAUUUAAGCUGCUC------------CCGCAGUGGCAGCU------GCAGCGACUU (((....)))..........(((---------......)))(((((((........((((((..------------(....)..))))))------.))))))).. ( -26.20) >consensus GUCAAUUGACACUUUUUGUUGCCAUUGCCUUCUCCUUUGGUGUCGCCGGCGAUUUAAGCUGCAGAUGCAACAGCAGCUGCAGCUGCAGCU______UCAGCGACUU (((....)))..........((((.............))))(((((.((.......((((((((.((((........)))).))))))))......)).))))).. (-27.90 = -28.79 + 0.89)

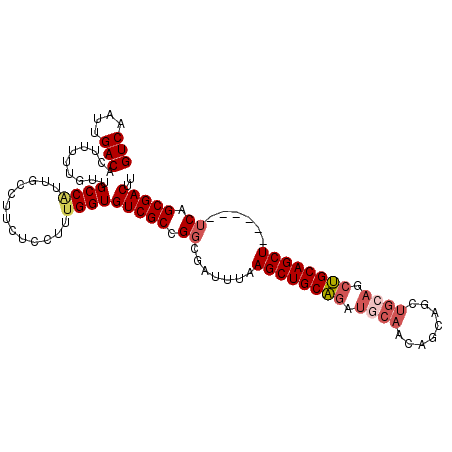
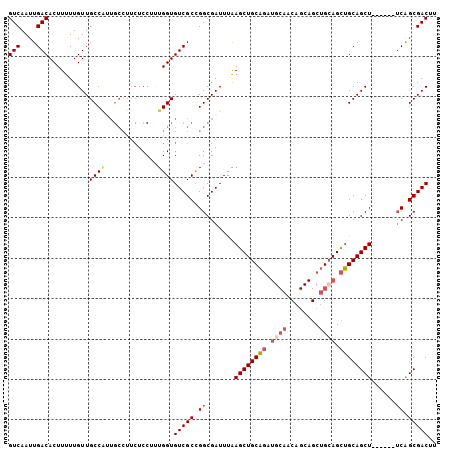
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:45 2006