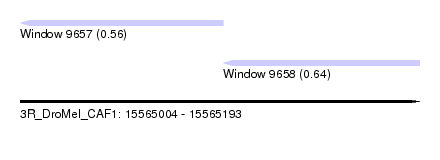
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,565,004 – 15,565,193 |
| Length | 189 |
| Max. P | 0.637303 |

| Location | 15,565,004 – 15,565,100 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 80.84 |
| Mean single sequence MFE | -29.00 |
| Consensus MFE | -18.08 |
| Energy contribution | -19.58 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555604 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
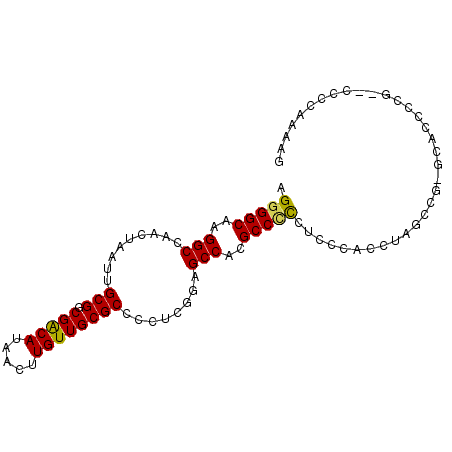
>3R_DroMel_CAF1 15565004 96 - 27905053 AGGCGUUCGCCACAUGCCUGUGUGUGUGUAGCGUGUGAUUUUGUGGCGCUUGCGUGUGUGUGUAAUAAACGCCGCAGAUAGACGGACGUACUAUAU ..(((((((.(((((((.............)))))))..(((((((((.(((((......)))))....)))))))))....)))))))....... ( -31.52) >DroSec_CAF1 90213 94 - 1 AGGCGUUCGCCACAUGCAUGUGUGUGUGUAGCGUGUGAUUUUGUGGCGCGU--GUGUGUGUGUAAUAAACGCCGCAGAUAGACGGACGUACUAUAU ..((((((((((((..((..(((.......)))..))....))))))..((--.((((.((((.....)))))))).))....))))))....... ( -32.90) >DroSim_CAF1 84198 94 - 1 AGGCGUUCGCCACAUGCAUGUGUGUGUGUAGCGUGUGAUUUUGUGGCGCGU--GUGUGUGUGUAAUAAACGCCGCAGAUAGACGGACGUACUAUAU ..((((((((((((..((..(((.......)))..))....))))))..((--.((((.((((.....)))))))).))....))))))....... ( -32.90) >DroYak_CAF1 88847 75 - 1 AGGCCUG-----------UGUAUGUGUACAGUGUGAU--UU-------UGUGG-GGUGUGUGUAAUAAACGCCGCAGAUAGACAGACGUACUAUAU .......-----------.(((((.((((.((.((..--((-------(((((-.((...........)).)))))))....)).))))))))))) ( -18.70) >consensus AGGCGUUCGCCACAUGCAUGUGUGUGUGUAGCGUGUGAUUUUGUGGCGCGU__GUGUGUGUGUAAUAAACGCCGCAGAUAGACGGACGUACUAUAU ..((((((((((((..(((..((.......))..)))....))))))........(((.((((.....)))))))........))))))....... (-18.08 = -19.58 + 1.50)

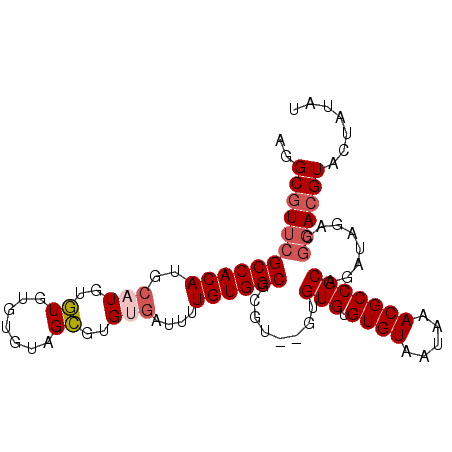
| Location | 15,565,100 – 15,565,193 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 81.13 |
| Mean single sequence MFE | -27.79 |
| Consensus MFE | -21.75 |
| Energy contribution | -21.53 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.637303 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15565100 93 - 27905053 AGGGGCAAGGCCAACUAAUUGCGGCGACAUAACUUGUUGCGCCCCUCGGAGCCACGCCCCCUCCCACCUAACCGUGCACCCCGCAU-CCAAAAG .(((((..((((........(((.(((((.....))))))))......).)))..))))).............((((.....))))-....... ( -26.74) >DroSec_CAF1 90307 93 - 1 AGGGGCAAGGCCAACUAAUUGCGGCGACAUAACUUGUUGCGCCCCUCGGAGCCACGCC-CCUCCCACUUAGCCGAGCACCCCGCGCCCCAAAAG .(((((..(((.........(((.(((((.....)))))))).....((((.......-.))))......)))..((.....)))))))..... ( -31.50) >DroSim_CAF1 84292 94 - 1 AGGGGCAAGGCCAACUAAUUGCGGCGACAUAACUUGUUGCGCCCCUCGGAGCCACGCCCCCUCCCACCUAGCCGAGCACCACGCGCCCCAAAAG .(((((..(((.........(((.(((((.....)))))))).....((((.........))))......)))..((.....)))))))..... ( -30.70) >DroEre_CAF1 79806 78 - 1 AGGGGCAAGGCCAACUAAUUGCGGCGACAUAACUUGUUGCGCCCCUCGCUGCCACGCCCCCUCC--------------UCCCG--CCCCCAAAG .(((((..(((.........(((.(((((.....))))))))........)))..)))))....--------------.....--......... ( -23.73) >DroYak_CAF1 88922 92 - 1 AGGGGCAAGGCCAACUAAUUGCGGCGACAUAACUUGUUGCGCCCCUCGGAGCCACGCCCUCUUCCUACUAGCCGUGCACCCCG--CCCCCAAAG .(((((..((((((....)))..((((((.....)))))))))....((.((.(((.(............).))))).))..)--))))..... ( -25.90) >DroAna_CAF1 90990 72 - 1 AGGGGCAAGGCCAACUAAUUGCGGCGGCAUAACUUGUUGCGCCCCUUGGAGCCACGCCUC-UGCCCCU------------------CCCCA--- (((((((((((............((((((.....))))))((((...)).))...)))).-)))))))------------------.....--- ( -28.20) >consensus AGGGGCAAGGCCAACUAAUUGCGGCGACAUAACUUGUUGCGCCCCUCGGAGCCACGCCCCCUCCCACCUAGCCG_GCACCCCG__CCCCAAAAG .(((((..(((.........(((.(((((.....))))))))........)))..))))).................................. (-21.75 = -21.53 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:38 2006