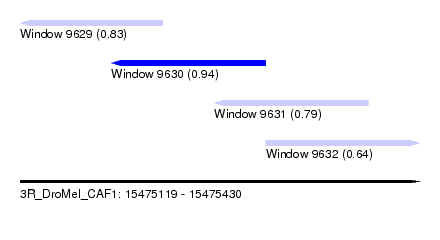
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,475,119 – 15,475,430 |
| Length | 311 |
| Max. P | 0.938141 |

| Location | 15,475,119 – 15,475,230 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.50 |
| Mean single sequence MFE | -28.30 |
| Consensus MFE | -25.01 |
| Energy contribution | -25.80 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15475119 111 - 27905053 UAAUAUAUGGGUCGGCAAUCCUUAAGUCGGAUGCACUCCUCAUCCGGUUCACACAGUUUAUCUAUUGAAAAGUAGAGAG-----UGUU----CAUAGUGUGUUCAUAAAUUGUUUUAAUC .......((..(((((.........)))(((((.......)))))))..)).((((((((((((((....))))))(((-----..(.----......)..))).))))))))....... ( -23.70) >DroSec_CAF1 7402 120 - 1 UUAUAUAUGUGUGGGCAAUCCUUAAGUUGGAUGCACUCCUCAUCCGAUUGACACAGUUUAUCUAUUGAAAAGUAGGGAAAUAAUUGUUAUAAGACAGUGUUUUCAUAAAUUGUAUUAAUC ......(((.(..((((.(((.......)))))).)..).)))..((((((.((((((((((((((....)))))(((((((.(((((....))))))))))))))))))))).)))))) ( -31.60) >DroSim_CAF1 2684 120 - 1 UCAUAUAUGUGUGGGCAAUCCUUAAGUCGGAUGCACUUCUCAUCCGGUUGACACAGUUUAUCUAUUGAAAAGUAGAGAAAUAAUUGUUAUAAGACAGUGUUUUCAUAAAUUGUAUUAAUC ......(((.(.(((((.(((.......)))))).)).).)))..((((((.((((((((((((((....))))))((((..((((((....))))))..)))).)))))))).)))))) ( -29.60) >consensus UAAUAUAUGUGUGGGCAAUCCUUAAGUCGGAUGCACUCCUCAUCCGGUUGACACAGUUUAUCUAUUGAAAAGUAGAGAAAUAAUUGUUAUAAGACAGUGUUUUCAUAAAUUGUAUUAAUC ......(((.(..((((.(((.......)))))).)..).)))..((((((.((((((((((((((....))))))((((..((((((....))))))..)))).)))))))).)))))) (-25.01 = -25.80 + 0.79)

| Location | 15,475,190 – 15,475,310 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.50 |
| Mean single sequence MFE | -30.71 |
| Consensus MFE | -17.59 |
| Energy contribution | -20.53 |
| Covariance contribution | 2.94 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15475190 120 - 27905053 CUCGAACACUCACGCCACACAUUCUUGUCUGCGAUAUAUUCACUUUUGCGGACCCCGACAAUCUGUUUCUCCUCACAUUGUAAUAUAUGGGUCGGCAAUCCUUAAGUCGGAUGCACUCCU ............(((...(((....)))..)))...(((((((((((((.(((((..(((((.((........)).))))).......))))).)))).....)))).)))))....... ( -25.00) >DroSec_CAF1 7482 120 - 1 AUCGAACAUUGUCGCCACAUAUUCUUGUCUGCAAUAUAUCUAAUUUUGCAGACCGCGACAAUCUUUUGCUCUUCAGAUCGUUAUAUAUGUGUGGGCAAUCCUUAAGUUGGAUGCACUCCU (((.(((((((((((...........((((((((...........)))))))).))))))))...((((((..((.((........)).)).)))))).......))).)))........ ( -30.60) >DroSim_CAF1 2764 120 - 1 AUCGAACAUUGUCGCCACAUAUUCUUGUCUGCAAUAUAUCUAAUUUUGCAGACCACGACAAUCUUUUGCUCUUCAGAUCGUCAUAUAUGUGUGGGCAAUCCUUAAGUCGGAUGCACUUCU ...(((........((((((((....((((((((...........))))))))...(((.((((..........)))).)))....))))))))(((.(((.......))))))..))). ( -31.10) >DroYak_CAF1 2455 120 - 1 UACGAACACUGGAGCCACAUCGCCUGUUGUUCAAAAUUUCCCUUUUUGUGGCUCUGGACAUUCUGUUUGUCUUCACAGCGUCAUAAACUUGUGGGCAAUCCUGCAGUCUGACGCACGUCU .((((((((..((((((((......(((......))).........))))))))..)......))))))).......((((((...(((.(..((....))..)))).))))))...... ( -36.16) >consensus AUCGAACACUGUCGCCACAUAUUCUUGUCUGCAAUAUAUCCAAUUUUGCAGACCACGACAAUCUGUUGCUCUUCACAUCGUCAUAUAUGUGUGGGCAAUCCUUAAGUCGGAUGCACUCCU ..............((((((((....((((((((...........))))))))...(((.(((((........))))).)))....))))))))(((.(((.......))))))...... (-17.59 = -20.53 + 2.94)


| Location | 15,475,270 – 15,475,390 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.78 |
| Mean single sequence MFE | -38.03 |
| Consensus MFE | -21.23 |
| Energy contribution | -24.86 |
| Covariance contribution | 3.63 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15475270 120 - 27905053 CUUCCCACAAUCCGGAUAGCUUGGCAAGGUGUUUGCCGGCUUGGGACAGCAGAUAUAAAUCCUUUUGUAACGUUUCCGGUCUCGAACACUCACGCCACACAUUCUUGUCUGCGAUAUAUU .........((((((((((..((((..((((((((((((..((..((((.((.........)).))))..))...)))))...)))))))...)))).......))))))).)))..... ( -30.40) >DroSec_CAF1 7562 120 - 1 CCGUCCACAAGCCGGAUAGCUUGGCAAGAUGGUUGCCGGCUUGGGACAGCAGAUAUAAAUCAUUUGGUUAAGAUUCUGGUAUCGAACAUUGUCGCCACAUAUUCUUGUCUGCAAUAUAUC ..((((.((((((((.(((((.........))))))))))))))))).(((((((..(((.((.((((...(((..((........))..))))))).)))))..)))))))........ ( -43.50) >DroSim_CAF1 2844 120 - 1 CCGUCCACAAGCCGGAUAGCUUGGCAAGAUGGUUGCCGGCUUGGGACAGCAGAUAUAACUCAUUUGGUUAAGAUUCUGGUAUCGAACAUUGUCGCCACAUAUUCUUGUCUGCAAUAUAUC ..((((.((((((((.(((((.........))))))))))))))))).(((((((.........((((...(((..((........))..)))))))........)))))))........ ( -41.53) >DroYak_CAF1 2535 117 - 1 UUGUCCGCAAGCCGAAAGUCUGGGCAAGAUGUCUGCGGGUUCGGGACAGCAGAUG---GUUCUAUGGUGAAGUUUCUGCUUACGAACACUGGAGCCACAUCGCCUGUUGUUCAAAAUUUC ((((((...((.(....).))))))))(((((((.((((((((....((((((.(---.(((......))).).))))))..))))).))).))..)))))................... ( -36.70) >consensus CCGUCCACAAGCCGGAUAGCUUGGCAAGAUGGUUGCCGGCUUGGGACAGCAGAUAUAAAUCAUUUGGUUAAGAUUCUGGUAUCGAACACUGUCGCCACAUAUUCUUGUCUGCAAUAUAUC ..((((.((((((((.(((((.........))))))))))))))))).(((((((.........((((...(((..((........))..)))))))........)))))))........ (-21.23 = -24.86 + 3.63)

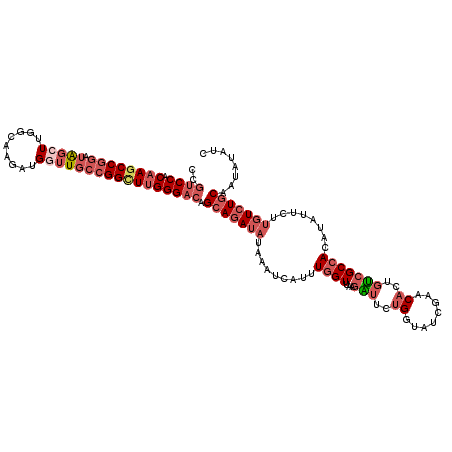
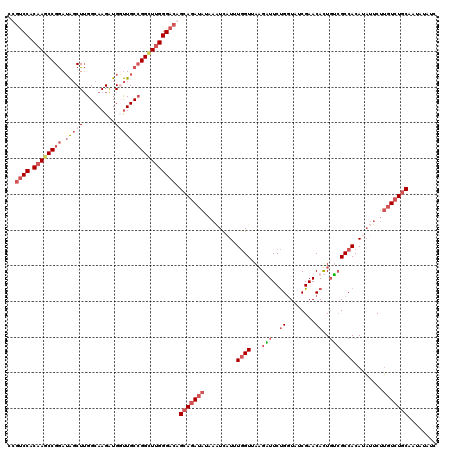
| Location | 15,475,310 – 15,475,430 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.86 |
| Mean single sequence MFE | -38.30 |
| Consensus MFE | -26.51 |
| Energy contribution | -28.20 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15475310 120 + 27905053 ACCGGAAACGUUACAAAAGGAUUUAUAUCUGCUGUCCCAAGCCGGCAAACACCUUGCCAAGCUAUCCGGAUUGUGGGAAGCCACAAACGACUAACCGUGUUAUAGGCGGCACGGAGCCGA .(((((...(..(((...((((....))))..)))..).(((.(((((.....)))))..))).))))).((((((....)))))).((.((..(((((((......))))))))).)). ( -40.00) >DroSec_CAF1 7602 120 + 1 ACCAGAAUCUUAACCAAAUGAUUUAUAUCUGCUGUCCCAAGCCGGCAACCAUCUUGCCAAGCUAUCCGGCUUGUGGACGGCCACAAUUGAUUUACCGUGUUAUGGGUGGCACGGAGCCGA ....(((((..........(((....))).((((((((((((((((((.....)))))..(.....))))))).))))))).......))))).((((((((....))))))))...... ( -40.90) >DroSim_CAF1 2884 120 + 1 ACCAGAAUCUUAACCAAAUGAGUUAUAUCUGCUGUCCCAAGCCGGCAACCAUCUUGCCAAGCUAUCCGGCUUGUGGACGGCCACAAUUGACUUACCGGGUUAUGGGUGGCACGGAGCCGA ......(((((((((...(((((((.....((((((((((((((((((.....)))))..(.....))))))).)))))))......)))))))...))))).))))(((.....))).. ( -45.30) >DroYak_CAF1 2575 117 + 1 AGCAGAAACUUCACCAUAGAAC---CAUCUGCUGUCCCGAACCCGCAGACAUCUUGCCCAGACUUUCGGCUUGCGGACAAAUGAAAAUGACUUUCCGGAUUAUGGGUGGCACGGAGCCGG ((((((...(((......))).---..))))))(..(((...((((((.......(((.........)))))))))...........((.((.((((.....)))).)))))))..)... ( -27.01) >consensus ACCAGAAACUUAACCAAAGGAUUUAUAUCUGCUGUCCCAAGCCGGCAAACAUCUUGCCAAGCUAUCCGGCUUGUGGACAGCCACAAAUGACUUACCGGGUUAUGGGUGGCACGGAGCCGA ..............................((((((((((((((((((.....)))))..(.....))))))).))))))).............((((((((....))))))))...... (-26.51 = -28.20 + 1.69)

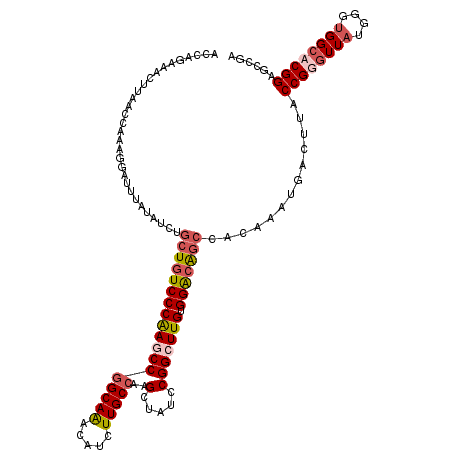
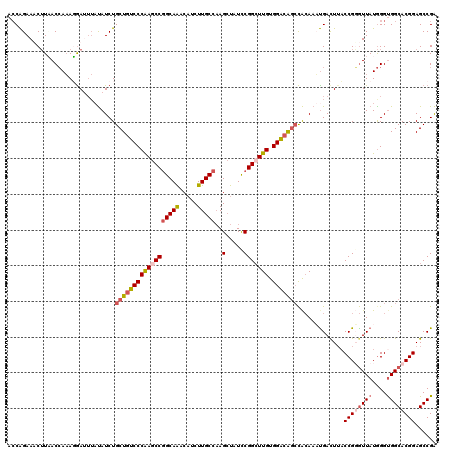
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:12 2006