
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,471,076 – 15,471,313 |
| Length | 237 |
| Max. P | 0.988174 |

| Location | 15,471,076 – 15,471,196 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.72 |
| Mean single sequence MFE | -52.83 |
| Consensus MFE | -24.26 |
| Energy contribution | -24.88 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.54 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.46 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15471076 120 - 27905053 GUAUGCCAAAAGCGGGCGUGGCAGCAGCCGUUCCAAAAGCCGGCGCCGCAGUGGAAGUGGCGCCAAAACCAGGCGUGGACGCAGGUGCUCCAAAAGCCGGCGUAGCAGCUGGUGCCCCAA ..(((((.......)))))((((.((((.(((......(((((((((((.......)))))))).......(((.((((.((....))))))...))))))..))).)))).)))).... ( -56.90) >DroSec_CAF1 1836 120 - 1 GUACGCCAAAAGCGGGCGUGGCAGCAGCCGUCCCAAGAGCCGGCGUCGCAGUGGAAGUGGCGCCAAAACCAGGCGUGGACGCAGGAGCUCCAAAAGCCGGCGUAGCAGCUGGUGCCCCAA ..(((((.......)))))((((.((((.((..(....(((((((((((.......)))))))).......(((.((((.((....))))))...)))))))..)).)))).)))).... ( -55.00) >DroSim_CAF1 1835 120 - 1 GUACGCCAAAAGCGGGCGUGGCAGCAGCCGUUCCAAAAGCCGGCGUCGCAGUGGAAGUGGCGCCAAAACCAGGCGUGGACGCAGGCGCUCCAAAAGCCGGCGUAGCAGCUGGUGCCCCAA ..(((((.......)))))((((.((((.(((......(((((((((((.......)))))))).......(((.((((((....)).))))...))))))..))).)))).)))).... ( -55.70) >DroEre_CAF1 1761 120 - 1 GUGCUCCAAAAGCAGCCGUAGCAGCUGGUGCUCCAAAGGCCGGAGUGGCCGCCGGAGCUGCAAACAAGGAAGUGGUGCCCAGAGCAGCACCGAAGGACGUCGUGGAGGUGGCUGCUCCAA .((((.....))))(((...(.(((....))).)...))).((((..((((((....(..(............(((((........)))))((......)))..).))))))..)))).. ( -46.50) >DroYak_CAF1 1833 99 - 1 UUGA------A---------------GUUGCGCCAAAGGCAGGCGUAGCAGUUGAAGUUGCGCCAAAGGCAGGCGUAGCAGCUGGUGCUCUAAAAGCCGGCGUAGCAGCUGGUGCUCCGA ....------.---------------((((((((....((.((((((((.......))))))))....)).(((.(((.(((....))))))...)))))))))))(((....))).... ( -42.30) >DroAna_CAF1 1374 120 - 1 CGGCUCCAAAAGCUGGUGCGGUUGUAGCUGCUCCAAAGGCUGGCGCAGCCGCCGCUGGUGCUCCAAAAGCGGACGUUGCAGCGGGAGCACCAAACGCCGGGGCGGCAGAUACUGCCCCGA (((((.....)))))..(((((....(((((.(((.....))).))))).)))))(((((((((....((..........)).))))))))).....((((((((......)))))))). ( -60.60) >consensus GUACGCCAAAAGCGGGCGUGGCAGCAGCCGCUCCAAAAGCCGGCGUCGCAGUGGAAGUGGCGCCAAAACCAGGCGUGGACGCAGGAGCUCCAAAAGCCGGCGUAGCAGCUGGUGCCCCAA .................(.((((.((((.((..(.......((((((((.......)))))))).......(((.(((...........)))...)))...)..)).)))).)))).).. (-24.26 = -24.88 + 0.62)

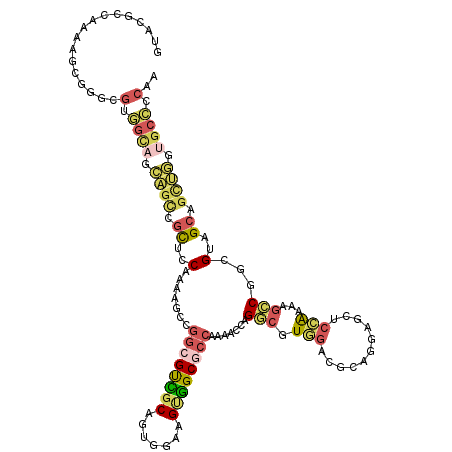
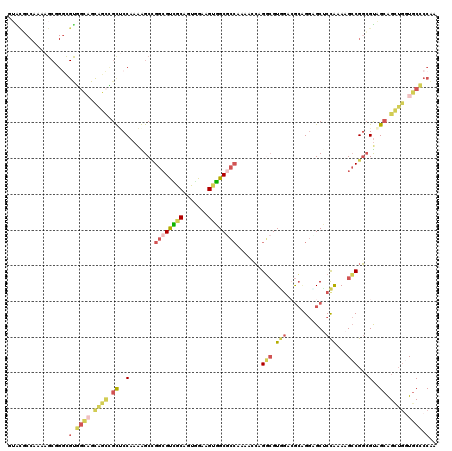
| Location | 15,471,116 – 15,471,233 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 66.67 |
| Mean single sequence MFE | -51.94 |
| Consensus MFE | -19.14 |
| Energy contribution | -21.12 |
| Covariance contribution | 1.98 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15471116 117 - 27905053 GUAGCAGCU---GGAGCUCCAAAAGCCGACGUUGCUGCUGGUAUGCCAAAAGCGGGCGUGGCAGCAGCCGUUCCAAAAGCCGGCGCCGCAGUGGAAGUGGCGCCAAAACCAGGCGUGGAC ......((.---...))((((...(((((((..(((((((.((((((.......)))))).))))))))))).........((((((((.......)))))))).......))).)))). ( -54.10) >DroSec_CAF1 1876 117 - 1 GUAGCAGCU---GGAGCUCCAAAAGCCGACGUUGCUGCUGGUACGCCAAAAGCGGGCGUGGCAGCAGCCGUCCCAAGAGCCGGCGUCGCAGUGGAAGUGGCGCCAAAACCAGGCGUGGAC ......((.---...))((((...(((((((..(((((((.((((((.......)))))).))))))))))).........((((((((.......)))))))).......))).)))). ( -55.50) >DroSim_CAF1 1875 117 - 1 GUGGCAGCU---GGAGCUCCAAAAGCCGACGUUGCUGCUGGUACGCCAAAAGCGGGCGUGGCAGCAGCCGUUCCAAAAGCCGGCGUCGCAGUGGAAGUGGCGCCAAAACCAGGCGUGGAC .((((...(---((....)))...))))(((..(((((((.((((((.......)))))).))))))))))((((...(((((((((((.......)))))))).......))).)))). ( -55.11) >DroEre_CAF1 1801 120 - 1 GCUGCAGUUGAAGUUGCGCCAAAAGCAGGCGUGGACGCAGGUGCUCCAAAAGCAGCCGUAGCAGCUGGUGCUCCAAAGGCCGGAGUGGCCGCCGGAGCUGCAAACAAGGAAGUGGUGCCC (((((...((...(..((((.......))))..)...))((((((.....)))).))...)))))....(((((...((((.....))))...))))).(((.((......))..))).. ( -45.70) >DroAna_CAF1 1414 117 - 1 GCAGCUGUA---GCGGCUCCGAAAGCUGGCGCGGCUGUAGCGGCUCCAAAAGCUGGUGCGGUUGUAGCUGCUCCAAAGGCUGGCGCAGCCGCCGCUGGUGCUCCAAAAGCGGACGUUGCA (((((....---((((((.((..((((((.(((((((((((.((.(((.....))).)).))))))))))).))...))))..)).))))))((((..((...))..))))...))))). ( -58.60) >DroPer_CAF1 1783 114 - 1 UGUGCCGUA---GGUGCUCCGAAUCCGGGCUGGGCCGUAGGUGCUCCAAAUCCUGGCUGCGCCGUGGGUGCACCAAACCCAGACUGUGCCGUGGG---UGCUCCGAAUCCUGGCUGGGCU .(..((...---))..).(((....)))(((.((((...(((((.(((.....)))..)))))...((.(((((.....((.....)).....))---))).)).......)))).))). ( -42.60) >consensus GUAGCAGCU___GGAGCUCCAAAAGCCGACGUGGCUGCAGGUACGCCAAAAGCGGGCGUGGCAGCAGCCGCUCCAAAAGCCGGCGUCGCAGUGGAAGUGGCGCCAAAACCAGGCGUGGAC ((((((((.......((.......))....))))))))...((((((.......((.(((((....))))).)).......((.(((((.......))))).)).......))))))... (-19.14 = -21.12 + 1.98)

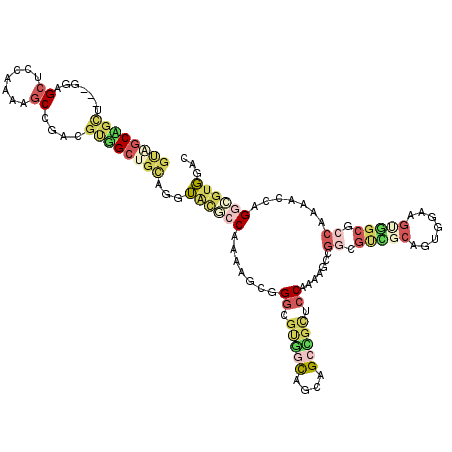
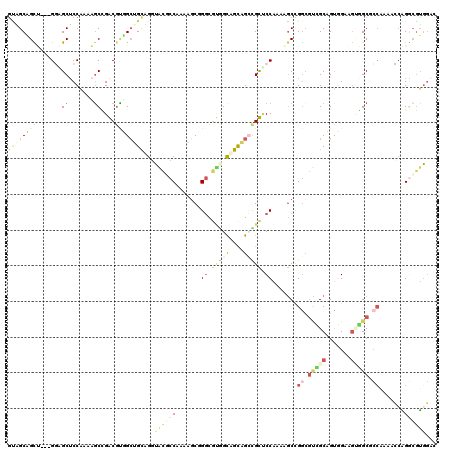
| Location | 15,471,156 – 15,471,273 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.68 |
| Mean single sequence MFE | -53.00 |
| Consensus MFE | -23.42 |
| Energy contribution | -23.90 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.85 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15471156 117 - 27905053 UCCGAAAGCGGGAGUGGCAGCUGCAGCUCCAAAAGCGGGCGUAGCAGCU---GGAGCUCCAAAAGCCGACGUUGCUGCUGGUAUGCCAAAAGCGGGCGUGGCAGCAGCCGUUCCAAAAGC .......((.(((((..(((((((.(((((....).))))...))))))---)..)))))....)).((((..(((((((.((((((.......)))))).)))))))))))........ ( -54.10) >DroSec_CAF1 1916 117 - 1 UCCAAAGGCGGGCGUGGCAGCUGGAGCUCCAAAAGCGGGCGUAGCAGCU---GGAGCUCCAAAAGCCGACGUUGCUGCUGGUACGCCAAAAGCGGGCGUGGCAGCAGCCGUCCCAAGAGC ......((((((((((((...((((((((((...((..((...)).)))---)))))))))...))).)))))(((((((.((((((.......)))))).)))))))))))........ ( -59.90) >DroSim_CAF1 1915 117 - 1 GCCAAAAGCGGGCGUGGCGGCGGUUGCUCCAAAAGCGGGCGUGGCAGCU---GGAGCUCCAAAAGCCGACGUUGCUGCUGGUACGCCAAAAGCGGGCGUGGCAGCAGCCGUUCCAAAAGC (((.......))).(((.(((((((((((((...((.((((((.(((((---((....)))..(((.......))))))).))))))....)).....))).)))))))))))))..... ( -52.10) >DroEre_CAF1 1841 120 - 1 UCCAAAGGCCGGCGUGGCUGCUGGUGCGCCAAAAGCAGGCGCUGCAGUUGAAGUUGCGCCAAAAGCAGGCGUGGACGCAGGUGCUCCAAAAGCAGCCGUAGCAGCUGGUGCUCCAAAGGC .......((((((.((((((((((.(((((....((.(((((.((.......)).)))))....))..(((....))).))))).))...)))))))).....))))))(((.....))) ( -50.70) >DroAna_CAF1 1454 117 - 1 UCCAAAGGCUGGUGCGGUUGUAGCAGAUCCAAAGGCUGGUGCAGCUGUA---GCGGCUCCGAAAGCUGGCGCGGCUGUAGCGGCUCCAAAAGCUGGUGCGGUUGUAGCUGCUCCAAAGGC .((...(((((.((((((((((.(((.((....))))).))))))))))---.)))))........(((.(((((((((((.((.(((.....))).)).))))))))))).)))..)). ( -54.90) >DroPer_CAF1 1820 117 - 1 UCCGAAUCCGGGCUGGGCCGUGGGAGCUCCAAACCCGGACUGUGCCGUA---GGUGCUCCGAAUCCGGGCUGGGCCGUAGGUGCUCCAAAUCCUGGCUGCGCCGUGGGUGCACCAAACCC ........(((.(..(.((((.(((((.((.....(((......)))..---)).))))).)...))).)..).)))..(((((.(((.....)))..)))))..((((.......)))) ( -46.30) >consensus UCCAAAGGCGGGCGUGGCAGCUGGAGCUCCAAAAGCGGGCGUAGCAGCU___GGAGCUCCAAAAGCCGACGUGGCUGCAGGUACGCCAAAAGCGGGCGUGGCAGCAGCCGCUCCAAAAGC ......((((..((((.(((((..((((((...(((..........)))...)))))).....))))).))))(((((((.(((.((.......)).))).)))))))))))........ (-23.42 = -23.90 + 0.48)
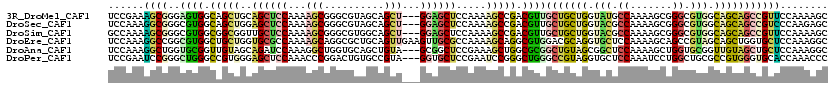


| Location | 15,471,196 – 15,471,313 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.92 |
| Mean single sequence MFE | -56.33 |
| Consensus MFE | -26.22 |
| Energy contribution | -25.15 |
| Covariance contribution | -1.07 |
| Combinations/Pair | 1.65 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15471196 117 - 27905053 UAGCGGCGGGUGCUCCAAAAGCGGGCGUGGCGGCGGGUGCUCCGAAAGCGGGAGUGGCAGCUGCAGCUCCAAAAGCGGGCGUAGCAGCU---GGAGCUCCAAAAGCCGACGUUGCUGCUG (((((((...((((.....))))((((((((....((.((((((...((....))..)((((((.(((((....).))))...))))))---))))).))....))).)))))))))))) ( -53.20) >DroSec_CAF1 1956 117 - 1 UGGCGGCGGCUGCUCCAAAAGCGGGCGUGGCGGCGGUUGCUCCAAAGGCGGGCGUGGCAGCUGGAGCUCCAAAAGCGGGCGUAGCAGCU---GGAGCUCCAAAAGCCGACGUUGCUGCUG .(((((((((((((.....))))(((((.((.(((.(((((.....))))).))).)).))((((((((((...((..((...)).)))---)))))))))...)))...))))))))). ( -57.50) >DroSim_CAF1 1955 117 - 1 UGGCGGCGGGUGCUCCAAAAGCGGGCGUGGCGGCGGUUGCGCCAAAAGCGGGCGUGGCGGCGGUUGCUCCAAAAGCGGGCGUGGCAGCU---GGAGCUCCAAAAGCCGACGUUGCUGCUG .((((((.((.(((((....((.((.(..((.((.((..((((.......))))..)).)).))..).))....)).(((......)))---))))).))...(((....))))))))). ( -56.40) >DroEre_CAF1 1881 120 - 1 CGGCAGCUGGCGCUCCAAAGGCCGGCGUGGCUGCUGGCGCUCCAAAGGCCGGCGUGGCUGCUGGUGCGCCAAAAGCAGGCGCUGCAGUUGAAGUUGCGCCAAAAGCAGGCGUGGACGCAG ..(((((((((.((....))))))))(((.((((((((((.(((..((((.....))))..))).)))))...))))).))))))...((...(..((((.......))))..)...)). ( -59.80) >DroYak_CAF1 1932 114 - 1 UGGAGGCGGGGGCUCCAAAAGCGGAAGUGGCAGCUGGAGCUCCAAAAGCCGGCGUGGCAGCCGCUGCUCCAAAAGCGGGCGUGGCAGU---AGCUGCUCCAAAAGCGGGCGC---CGCAG ....(((.(((((((((...((....))......)))))))))....(((.((.(((.(((.(((((((((...(....).))).)))---))).))))))...)).)))))---).... ( -57.80) >DroAna_CAF1 1494 117 - 1 UGGAGGUAGCAGCUCCGAACGCUGGUGCAGCUGUAGCUGCUCCAAAGGCUGGUGCGGUUGUAGCAGAUCCAAAGGCUGGUGCAGCUGUA---GCGGCUCCGAAAGCUGGCGCGGCUGUAG ........((((((.((...(((((.(((((....))))).))...(((((.((((((((((.(((.((....))))).))))))))))---.))))).....)))...)).)))))).. ( -53.30) >consensus UGGCGGCGGGUGCUCCAAAAGCGGGCGUGGCGGCGGGUGCUCCAAAAGCGGGCGUGGCAGCUGCAGCUCCAAAAGCGGGCGUAGCAGCU___GGAGCUCCAAAAGCCGACGUUGCUGCAG (((.(((....)))))).........((((((((....(((.....))).(((.(((.(((.((((((((.......))...)))))).......))))))...)))...)))))))).. (-26.22 = -25.15 + -1.07)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:08 2006