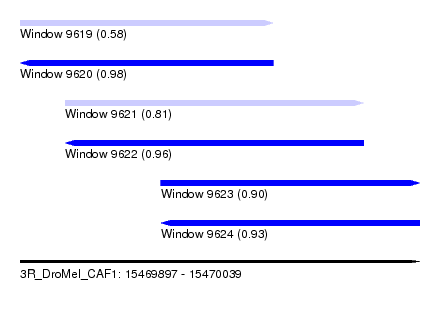
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,469,897 – 15,470,039 |
| Length | 142 |
| Max. P | 0.980911 |

| Location | 15,469,897 – 15,469,987 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 76.59 |
| Mean single sequence MFE | -24.61 |
| Consensus MFE | -15.94 |
| Energy contribution | -16.13 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
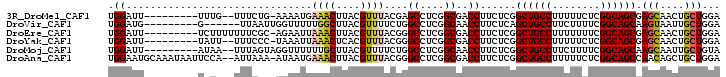
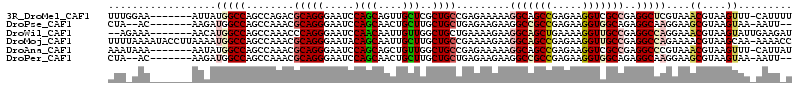
>3R_DroMel_CAF1 15469897 90 + 27905053 UGGAUU---------UUUG--UUUCUG-AAAAUGAAACUUACGUUUACGAGCCUCGGCGACCUUCUCGGCUGCCUUUUUCUCGGCAGCGAGCAACUGCUGGA .((...---------...(--((((..-.....)))))...((....))..))((((((.....((((.(((((........)))))))))....)))))). ( -25.00) >DroVir_CAF1 466 87 + 1 UGGAUG---------G------UUAAUUGGUUUUUGGCUUACGUUUUCUGGCCUCGGCAACCUUCUCAGCUGCCUUCUUUUCGGCAGCAAGUAAUUGCUGGA ......---------.------.............((((..........))))(((((((....((..((((((........)))))).))...))))))). ( -22.70) >DroEre_CAF1 672 92 + 1 UGGAUU---------UCUUUUUUUCGC-AGAAUUAAACUUACGUUUACGGGCCUCGGCGACCUUCUCGGCUGCCUUUUUUUCGGCAGCGAGCAACUGCUGGA ......---------..........((-((...(((((....))))).((((....))..))..((((.(((((........)))))))))...)))).... ( -23.40) >DroYak_CAF1 671 90 + 1 UGGAUU---------UAUU--UUUCCC-UAAAUUAAACUCACGUUUACGGGCCUCGGCGACCUUCUCGGCUGCCUUUUUCUCGGCAGCGAGCAACUGCUGGA ......---------....--...(((-(((((.........))))).)))..((((((.....((((.(((((........)))))))))....)))))). ( -23.40) >DroMoj_CAF1 471 91 + 1 UGGAUU---------AUAA--UUUAGUAGGUUUUUUGCUUACGUUUUCUGGCCUCGGCAACCUUCUCGGCUGCCUUCUUUUCGGCAGCAAGCAAUUGCUGUA .((...---------....--....((((((.....)))))).........))..((((.((.....)).)))).........(((((((....))))))). ( -25.35) >DroAna_CAF1 603 99 + 1 UGGAAUGCAAAUAAUUCCA--AUUAAA-AUAAUGAAACUUACGUUUACGGGCCUCGGCGACCUUCUCGGCUGCCUUUUUCUCGGCAGCCAACAGCUGCUGGA ((((((.......))))))--......-.............((....))..((.((((((.....))(((((((........)))))))....))))..)). ( -27.80) >consensus UGGAUU_________UCUA__UUUAAC_AGAAUUAAACUUACGUUUACGGGCCUCGGCGACCUUCUCGGCUGCCUUUUUCUCGGCAGCGAGCAACUGCUGGA .((...............................((((....))))....((....))..))......((((((........)))))).(((....)))... (-15.94 = -16.13 + 0.20)
| Location | 15,469,897 – 15,469,987 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 76.59 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -18.72 |
| Energy contribution | -18.00 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15469897 90 - 27905053 UCCAGCAGUUGCUCGCUGCCGAGAAAAAGGCAGCCGAGAAGGUCGCCGAGGCUCGUAAACGUAAGUUUCAUUUU-CAGAAA--CAAA---------AAUCCA ....(((((.....)))))((((.....(((.(((.....))).)))....)))).........(((((.....-..))))--)...---------...... ( -24.60) >DroVir_CAF1 466 87 - 1 UCCAGCAAUUACUUGCUGCCGAAAAGAAGGCAGCUGAGAAGGUUGCCGAGGCCAGAAAACGUAAGCCAAAAACCAAUUAA------C---------CAUCCA ....((((((.(((((((((........)))))).)))..))))))...(((............))).............------.---------...... ( -20.90) >DroEre_CAF1 672 92 - 1 UCCAGCAGUUGCUCGCUGCCGAAAAAAAGGCAGCCGAGAAGGUCGCCGAGGCCCGUAAACGUAAGUUUAAUUCU-GCGAAAAAAAGA---------AAUCCA ....((((...(((((((((........))))).))))..((((.....))))..(((((....)))))...))-))..........---------...... ( -29.80) >DroYak_CAF1 671 90 - 1 UCCAGCAGUUGCUCGCUGCCGAGAAAAAGGCAGCCGAGAAGGUCGCCGAGGCCCGUAAACGUGAGUUUAAUUUA-GGGAAA--AAUA---------AAUCCA (((........(((((((((........))))).))))..((((.....))))..(((((....))))).....-.)))..--....---------...... ( -28.30) >DroMoj_CAF1 471 91 - 1 UACAGCAAUUGCUUGCUGCCGAAAAGAAGGCAGCCGAGAAGGUUGCCGAGGCCAGAAAACGUAAGCAAAAAACCUACUAAA--UUAU---------AAUCCA ........((((((((.(((........(((((((.....)))))))..)))..(....))))))))).............--....---------...... ( -23.80) >DroAna_CAF1 603 99 - 1 UCCAGCAGCUGUUGGCUGCCGAGAAAAAGGCAGCCGAGAAGGUCGCCGAGGCCCGUAAACGUAAGUUUCAUUAU-UUUAAU--UGGAAUUAUUUGCAUUCCA (((((....(.(((((((((........))))))))).).((((.....)))).(.((((....))))).....-.....)--))))............... ( -29.80) >consensus UCCAGCAGUUGCUCGCUGCCGAAAAAAAGGCAGCCGAGAAGGUCGCCGAGGCCCGUAAACGUAAGUUUAAUUCU_ACGAAA__AAAA_________AAUCCA ....((.....(((((((((........)))))).)))..((((.....))))...........)).................................... (-18.72 = -18.00 + -0.72)

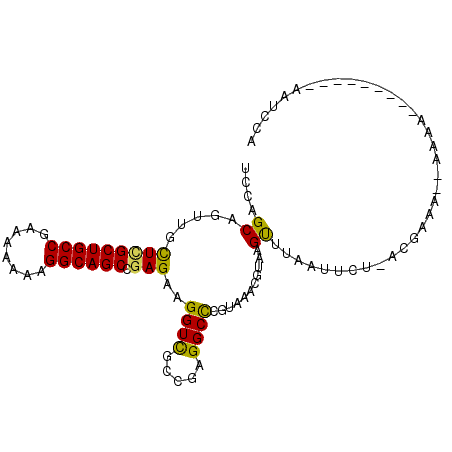
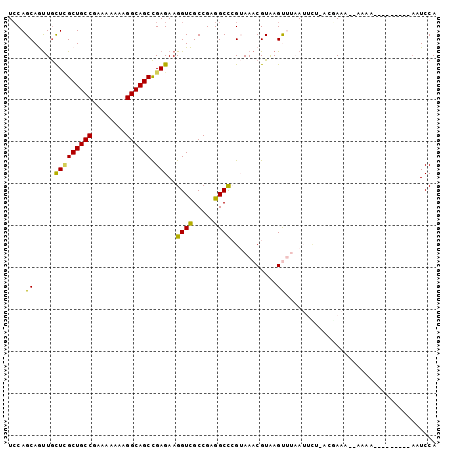
| Location | 15,469,913 – 15,470,019 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.47 |
| Mean single sequence MFE | -28.35 |
| Consensus MFE | -19.85 |
| Energy contribution | -19.93 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15469913 106 + 27905053 AAAAUG-AAACUUACGUUUACGAGCCUCGGCGACCUUCUCGGCUGCCUUUUUCUCGGCAGCGAGCAACUGCUGGAUUCCCUGCGUCUGGCUGGCCAUAAU-------UUCCAAA ...(((-((((....)))).(.((((..(((......((((.(((((........))))))))).....((.((....)).))))).)))).).)))...-------....... ( -28.90) >DroPse_CAF1 548 102 + 1 --AAUU-UUACUUACGCUUCCUUGCCUCUGCCACCUUCUCGGCGGCCUUCUUCUCAGCAGCAAGCAGUUGCUGGAUUCCCUGCGUUUGGCUGGCCAUCUU-------GU--UAG --....-...(..((((............(((.((.....)).))).....((.(((((((.....)))))))))......))))..).(((((......-------))--))) ( -25.70) >DroWil_CAF1 1893 105 + 1 AUCUUCAAUACUUACGUUUCCUGGCCUCGGCAACCUUUUCAGCUGCCUUCUUUUCAGCAGCCAACAAUUGUUGGAUUCCCUGGGUUUGGCUGGCCAUGUU-------UUUCU-- .....................(((((..(((.((((.....((((.........))))..(((((....))))).......))))...))))))))....-------.....-- ( -24.60) >DroMoj_CAF1 488 113 + 1 GGUUUU-UUGCUUACGUUUUCUGGCCUCGGCAACCUUCUCGGCUGCCUUCUUUUCGGCAGCAAGCAAUUGCUGUAUUCCCUGCGUUUGGCUGGCCAUUUUAAGGUAUUUUAAAA ((((..-..(((.((((.....((....((((.((.....)).)))).........(((((((....)))))))....)).))))..))).))))................... ( -30.40) >DroAna_CAF1 628 106 + 1 AUAAUG-AAACUUACGUUUACGGGCCUCGGCGACCUUCUCGGCUGCCUUUUUCUCGGCAGCCAACAGCUGCUGGAUUCCCUGCGUUUGGCUGGCCAUAUU-------UUUAUUU ..((((-(((....((....))((((..(((.........(((((((........)))))))..(((.(((.((....)).))).))))))))))....)-------)))))). ( -34.80) >DroPer_CAF1 541 102 + 1 --AAUU-UUACUUACGCUUCCUUGCCUCUGCCACCUUCUCGGCGGCCUUCUUCUCAGCAGCAAGCAGUUGCUGGAUUCCCUGCGUUUGGCUGGCCAUCUU-------GU--UAG --....-...(..((((............(((.((.....)).))).....((.(((((((.....)))))))))......))))..).(((((......-------))--))) ( -25.70) >consensus A_AAUU_AUACUUACGUUUCCUGGCCUCGGCAACCUUCUCGGCUGCCUUCUUCUCAGCAGCAAGCAAUUGCUGGAUUCCCUGCGUUUGGCUGGCCAUAUU_______UUU_UAA ....................((((((..((((.((.....)).))))......(((((((.......))))))).............))))))..................... (-19.85 = -19.93 + 0.08)
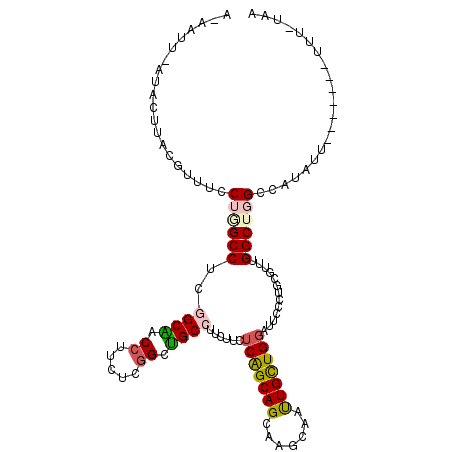
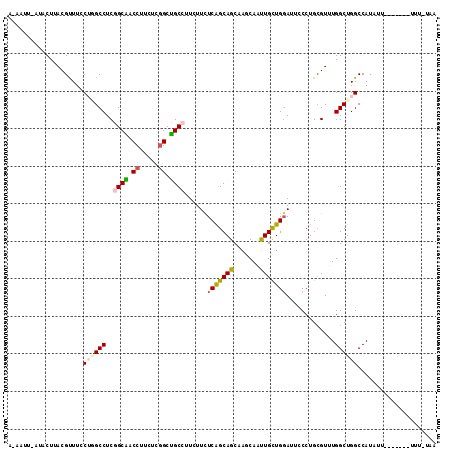
| Location | 15,469,913 – 15,470,019 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.47 |
| Mean single sequence MFE | -31.63 |
| Consensus MFE | -22.85 |
| Energy contribution | -23.55 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15469913 106 - 27905053 UUUGGAA-------AUUAUGGCCAGCCAGACGCAGGGAAUCCAGCAGUUGCUCGCUGCCGAGAAAAAGGCAGCCGAGAAGGUCGCCGAGGCUCGUAAACGUAAGUUU-CAUUUU ..(((((-------.(((((((.((((.(((((.((....)).)).....(((((((((........))))).))))...))).....)))).))...))))).)))-)).... ( -33.20) >DroPse_CAF1 548 102 - 1 CUA--AC-------AAGAUGGCCAGCCAAACGCAGGGAAUCCAGCAACUGCUUGCUGCUGAGAAGAAGGCCGCCGAGAAGGUGGCAGAGGCAAGGAAGCGUAAGUAA-AAUU-- ...--..-------....(((....))).((((.((....))........((((((.((....))...((((((.....))))))...))))))...))))......-....-- ( -28.80) >DroWil_CAF1 1893 105 - 1 --AGAAA-------AACAUGGCCAGCCAAACCCAGGGAAUCCAACAAUUGUUGGCUGCUGAAAAGAAGGCAGCUGAAAAGGUUGCCGAGGCCAGGAAACGUAAGUAUUGAAGAU --.....-------.((.(((((.........(((.(...(((((....))))).).))).......(((((((.....)))))))..)))))(....)))............. ( -30.80) >DroMoj_CAF1 488 113 - 1 UUUUAAAAUACCUUAAAAUGGCCAGCCAAACGCAGGGAAUACAGCAAUUGCUUGCUGCCGAAAAGAAGGCAGCCGAGAAGGUUGCCGAGGCCAGAAAACGUAAGCAA-AAAACC ...........((((...(((((........(((((.(((......))).)))))............(((((((.....)))))))..))))).......))))...-...... ( -32.20) >DroAna_CAF1 628 106 - 1 AAAUAAA-------AAUAUGGCCAGCCAAACGCAGGGAAUCCAGCAGCUGUUGGCUGCCGAGAAAAAGGCAGCCGAGAAGGUCGCCGAGGCCCGUAAACGUAAGUUU-CAUUAU .......-------....(((....)))......(((..((..((.(((.(((((((((........)))))))))...))).)).))..)))(.((((....))))-)..... ( -36.00) >DroPer_CAF1 541 102 - 1 CUA--AC-------AAGAUGGCCAGCCAAACGCAGGGAAUCCAGCAACUGCUUGCUGCUGAGAAGAAGGCCGCCGAGAAGGUGGCAGAGGCAAGGAAGCGUAAGUAA-AAUU-- ...--..-------....(((....))).((((.((....))........((((((.((....))...((((((.....))))))...))))))...))))......-....-- ( -28.80) >consensus CUA_AAA_______AAGAUGGCCAGCCAAACGCAGGGAAUCCAGCAACUGCUUGCUGCCGAGAAGAAGGCAGCCGAGAAGGUCGCCGAGGCCAGGAAACGUAAGUAA_AAUU_U ..................(((((........(((((.....)(((....)))..)))).........(((((((.....)))))))..)))))....((....))......... (-22.85 = -23.55 + 0.70)
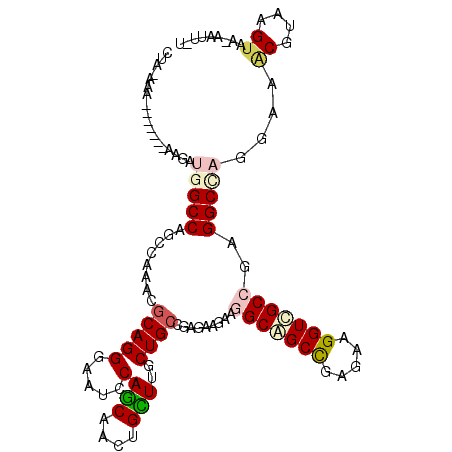
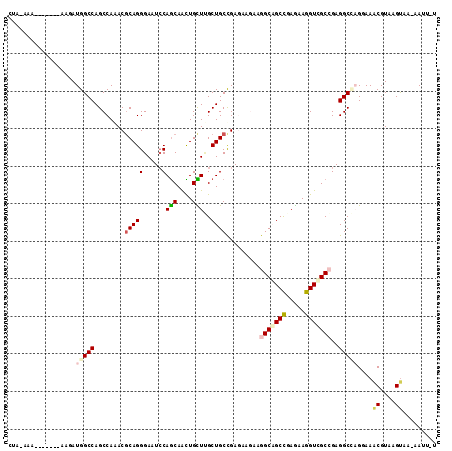
| Location | 15,469,947 – 15,470,039 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 75.28 |
| Mean single sequence MFE | -25.28 |
| Consensus MFE | -15.29 |
| Energy contribution | -15.15 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
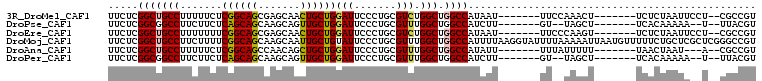
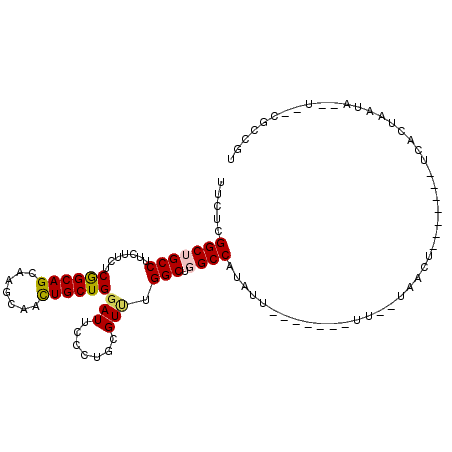
>3R_DroMel_CAF1 15469947 92 + 27905053 UUCUCGGCUGCCUUUUUCUCGGCAGCGAGCAACUGCUGGAUUCCCUGCGUCUGGCUGGCCAUAAU-------UUCCAAACU-------UCUCUAAUUCCU--CGCCGU ....((((((((........))))((.(((...(((.((....)).)))....))).))......-------.........-------............--.)))). ( -23.90) >DroPse_CAF1 580 88 + 1 UUCUCGGCGGCCUUCUUCUCAGCAGCAAGCAGUUGCUGGAUUCCCUGCGUUUGGCUGGCCAUCUU-------GU--UAGCU-------UCACAAAAA--U--UUACGU ....((((((......((.(((((((.....)))))))))....))))((..(((((((......-------))--)))))-------..)).....--.--...)). ( -25.30) >DroEre_CAF1 724 92 + 1 UUCUCGGCUGCCUUUUUUUCGGCAGCGAGCAACUGCUGGAUUCCCUGCGUCUGGCUGGCCAUAAU-------UUCCCAAGU-------UCUCUAAUUCCU--CGCCGU ....((((((((........))))))(((.((((((..(((.......)))..))(((.......-------...))))))-------))))........--...)). ( -24.40) >DroMoj_CAF1 522 108 + 1 UUCUCGGCUGCCUUCUUUUCGGCAGCAAGCAAUUGCUGUAUUCCCUGCGUUUGGCUGGCCAUUUUAAGGUAUUUUAAAAAUUAAUGUUUUUCUGCUCGCUCGGGCCGU ....((((.............(((((((....)))))))...((..(((...(((..(((.......))).....((((((....))))))..))))))..)))))). ( -27.40) >DroAna_CAF1 662 89 + 1 UUCUCGGCUGCCUUUUUCUCGGCAGCCAACAGCUGCUGGAUUCCCUGCGUUUGGCUGGCCAUAUU-------UUUAUUUUU-------UAACUAAU---A--CGCCGU .....(((((((........)))))))..(((.(((.((....)).))).)))...(((..((((-------.(((.....-------)))..)))---)--.))).. ( -25.40) >DroPer_CAF1 573 88 + 1 UUCUCGGCGGCCUUCUUCUCAGCAGCAAGCAGUUGCUGGAUUCCCUGCGUUUGGCUGGCCAUCUU-------GU--UAGCU-------UCACAAAAA--U--UUACGU ....((((((......((.(((((((.....)))))))))....))))((..(((((((......-------))--)))))-------..)).....--.--...)). ( -25.30) >consensus UUCUCGGCUGCCUUCUUCUCGGCAGCAAGCAACUGCUGGAUUCCCUGCGUUUGGCUGGCCAUAUU_______UU__UAACU_______UCACUAAUA__U__CGCCGU .....(((((((.......((((((.......))))))(((.......))).))).))))................................................ (-15.29 = -15.15 + -0.14)

| Location | 15,469,947 – 15,470,039 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 75.28 |
| Mean single sequence MFE | -25.82 |
| Consensus MFE | -15.91 |
| Energy contribution | -16.05 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
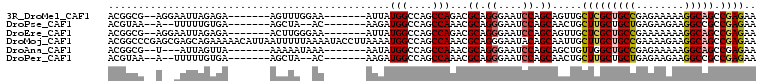
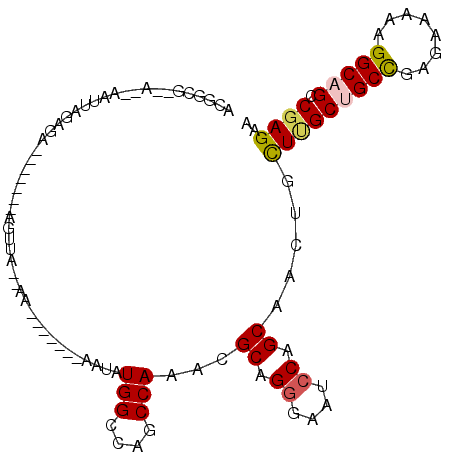
>3R_DroMel_CAF1 15469947 92 - 27905053 ACGGCG--AGGAAUUAGAGA-------AGUUUGGAA-------AUUAUGGCCAGCCAGACGCAGGGAAUCCAGCAGUUGCUCGCUGCCGAGAAAAAGGCAGCCGAGAA .(((((--((.((((.....-------.((((((..-------...........))))))((.((....)).)))))).)))..((((........)))))))).... ( -29.22) >DroPse_CAF1 580 88 - 1 ACGUAA--A--UUUUUGUGA-------AGCUA--AC-------AAGAUGGCCAGCCAAACGCAGGGAAUCCAGCAACUGCUUGCUGCUGAGAAGAAGGCCGCCGAGAA .((...--.--.((((((..-------.....--))-------)))).((((.((.....))......(((((((.(.....).))))).))....))))..)).... ( -20.30) >DroEre_CAF1 724 92 - 1 ACGGCG--AGGAAUUAGAGA-------ACUUGGGAA-------AUUAUGGCCAGCCAGACGCAGGGAAUCCAGCAGUUGCUCGCUGCCGAAAAAAAGGCAGCCGAGAA .(((((--((.((((.((..-------.((((.(..-------....(((....)))..).))))...))....)))).)))..((((........)))))))).... ( -26.90) >DroMoj_CAF1 522 108 - 1 ACGGCCCGAGCGAGCAGAAAAACAUUAAUUUUUAAAAUACCUUAAAAUGGCCAGCCAAACGCAGGGAAUACAGCAAUUGCUUGCUGCCGAAAAGAAGGCAGCCGAGAA .((((...((((((((((((((......)))))......(((.....(((....))).....)))...........)))))))))(((........))).)))).... ( -29.50) >DroAna_CAF1 662 89 - 1 ACGGCG--U---AUUAGUUA-------AAAAAUAAA-------AAUAUGGCCAGCCAAACGCAGGGAAUCCAGCAGCUGUUGGCUGCCGAGAAAAAGGCAGCCGAGAA .((((.--.---........-------.........-------....(((....)))...((.((....)).)).))))(((((((((........)))))))))... ( -28.70) >DroPer_CAF1 573 88 - 1 ACGUAA--A--UUUUUGUGA-------AGCUA--AC-------AAGAUGGCCAGCCAAACGCAGGGAAUCCAGCAACUGCUUGCUGCUGAGAAGAAGGCCGCCGAGAA .((...--.--.((((((..-------.....--))-------)))).((((.((.....))......(((((((.(.....).))))).))....))))..)).... ( -20.30) >consensus ACGGCG__A__AAUUAGAGA_______AGUUA__AA_______AAUAUGGCCAGCCAAACGCAGGGAAUCCAGCAACUGCUUGCUGCCGAGAAAAAGGCAGCCGAGAA ...............................................(((....)))...((.((....)).)).....(((((((((........))))).)))).. (-15.91 = -16.05 + 0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:07:04 2006