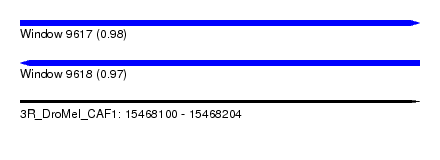
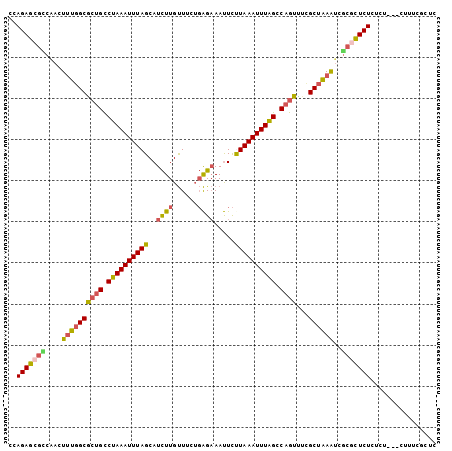
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,468,100 – 15,468,204 |
| Length | 104 |
| Max. P | 0.978413 |

| Location | 15,468,100 – 15,468,204 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 76.87 |
| Mean single sequence MFE | -27.33 |
| Consensus MFE | -22.02 |
| Energy contribution | -22.05 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.32 |
| Mean z-score | -3.45 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15468100 104 + 27905053 CCAGAGCGCCAACUUUGGCACUGCCUAAAUUUAGCAUCUUGUUUCUGAGAAAUUCCUAAAUUUAGUCAGUUUCGCUAAAUCGCAUUCUCUCUUCUUUUUCGCUC ...((((((((....))))((((.((((((((((..((((......)))).....)))))))))).))))...((......)).................)))) ( -26.10) >DroPse_CAF1 26261 90 + 1 ----AGACGCUUCUUCGGCGCCGUCUAAAUUUAGCAUUUUGCUUAGUGGCAACUUUUAAAUUUGGCCAAUC-GGCGGAGUCUUGCU---------GCUUCCCCC ----(((((((.....)))((((.((((((((((.(..(((((....)))))..))))))))))).....)-)))...))))....---------......... ( -22.90) >DroSec_CAF1 23604 104 + 1 CCAGAGCGCCAACUUUGGCACUGCCUAAAUUUAGCAUCUUGUUUCUGAGAAAUUCCUAAAUUUAGCCAGUUUCGCUAAAUCGCGCUCUCUCUUCUCCUUCGCUC ..(((((((....((((((((((.((((((((((..((((......)))).....)))))))))).))))...))))))..)))))))................ ( -30.80) >DroSim_CAF1 25006 104 + 1 CCAGAGCGCCAACUUUGGCGCUGCCUAAAUUUAGCAUCUUGUUUCUGAGAAAUUCUUAAAUUUAGCCAGUUUCGCUAAAUCGCGCUCUCUCUUCUCUUCCGCUC ..(((((((....((((((((((.((((((((((..((((......)))).....)))))))))).)))...)))))))..)))))))................ ( -29.30) >DroEre_CAF1 25269 101 + 1 CCAGAGCGUCGACUCUGGCGCUGCCUAAAUUUAGCAUCUUGUUGAUGAGAAAUUCUUAAAUUUAGCCAGUUUUGCUAAAUUGCGCUCUCUCU---CUUCCUCUC ..((((((.(((...((((((((.(((((((((.((((.....))))((.....))))))))))).))))...))))..)))))))))....---......... ( -27.20) >DroYak_CAF1 24618 91 + 1 CCAGAGCGGCAACUUUGGCGCUGCCUAAAUUUAGCAUCUUGUUGCUGAGAAAUUCUUAAAUUUAGUCAGUUUUGCUAAAUCGCGCUCUC-------------UC ..((((((.(...((((((((((.((((((((((((......))))(((.....))))))))))).))))...))))))..))))))).-------------.. ( -27.70) >consensus CCAGAGCGCCAACUUUGGCGCUGCCUAAAUUUAGCAUCUUGUUUCUGAGAAAUUCUUAAAUUUAGCCAGUUUCGCUAAAUCGCGCUCUCUCU___CUUUCGCUC ..(((((((....((((((((((.((((((((((..((((......)))).....)))))))))).))))...))))))..)))))))................ (-22.02 = -22.05 + 0.03)


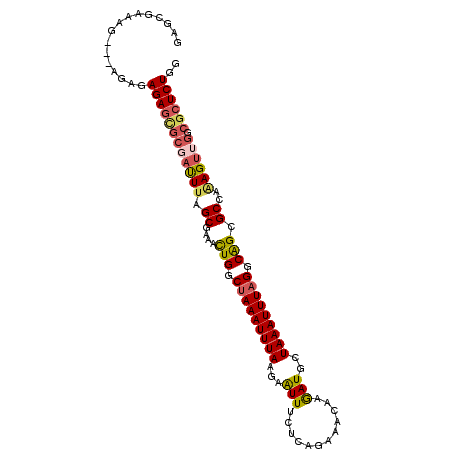
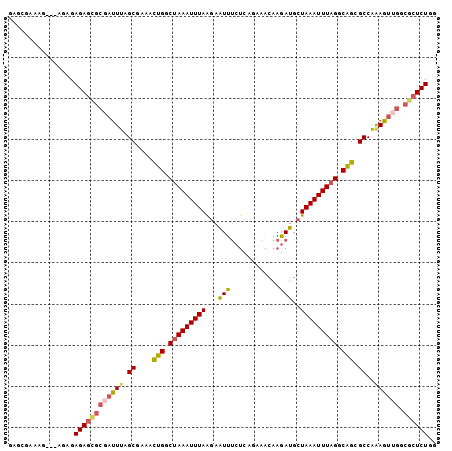
| Location | 15,468,100 – 15,468,204 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 76.87 |
| Mean single sequence MFE | -27.27 |
| Consensus MFE | -16.98 |
| Energy contribution | -17.65 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973916 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15468100 104 - 27905053 GAGCGAAAAAGAAGAGAGAAUGCGAUUUAGCGAAACUGACUAAAUUUAGGAAUUUCUCAGAAACAAGAUGCUAAAUUUAGGCAGUGCCAAAGUUGGCGCUCUGG ((((................(((......)))..((((.((((((((((.(...(((........)))).)))))))))).))))((((....))))))))... ( -27.40) >DroPse_CAF1 26261 90 - 1 GGGGGAAGC---------AGCAAGACUCCGCC-GAUUGGCCAAAUUUAAAAGUUGCCACUAAGCAAAAUGCUAAAUUUAGACGGCGCCGAAGAAGCGUCU---- ((.(((..(---------.....)..))).))-...((((..((((....))))))))...(((.....)))..........(((((.......))))).---- ( -19.60) >DroSec_CAF1 23604 104 - 1 GAGCGAAGGAGAAGAGAGAGCGCGAUUUAGCGAAACUGGCUAAAUUUAGGAAUUUCUCAGAAACAAGAUGCUAAAUUUAGGCAGUGCCAAAGUUGGCGCUCUGG ((((................(((......)))..((((.((((((((((.(...(((........)))).)))))))))).))))((((....))))))))... ( -31.60) >DroSim_CAF1 25006 104 - 1 GAGCGGAAGAGAAGAGAGAGCGCGAUUUAGCGAAACUGGCUAAAUUUAAGAAUUUCUCAGAAACAAGAUGCUAAAUUUAGGCAGCGCCAAAGUUGGCGCUCUGG ...(....).......((((((((((((.(((...(((.(((((((((.(....(((........)))..)))))))))).))))))..))))).))))))).. ( -30.00) >DroEre_CAF1 25269 101 - 1 GAGAGGAAG---AGAGAGAGCGCAAUUUAGCAAAACUGGCUAAAUUUAAGAAUUUCUCAUCAACAAGAUGCUAAAUUUAGGCAGCGCCAGAGUCGACGCUCUGG ..(..((.(---(((((...(..((((((((.......))))))))...)..)))))).))..)..(.((((.......)))).).(((((((....))))))) ( -27.60) >DroYak_CAF1 24618 91 - 1 GA-------------GAGAGCGCGAUUUAGCAAAACUGACUAAAUUUAAGAAUUUCUCAGCAACAAGAUGCUAAAUUUAGGCAGCGCCAAAGUUGCCGCUCUGG ..-------------.(((((..(((((((((...((((..(((((....))))).))))........)))))))))..((((((......))))))))))).. ( -27.40) >consensus GAGCGAAAG___AGAGAGAGCGCGAUUUAGCGAAACUGGCUAAAUUUAAGAAUUUCUCAGAAACAAGAUGCUAAAUUUAGGCAGCGCCAAAGUUGGCGCUCUGG ................((((((((((((.((....(((.(((((((((...(((............)))..))))))))).))).))..)))))).)))))).. (-16.98 = -17.65 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:58 2006