| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,191,627 – 2,191,730 |
| Length | 103 |
| Max. P | 0.633057 |

| Location | 2,191,627 – 2,191,730 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 76.39 |
| Mean single sequence MFE | -22.44 |
| Consensus MFE | -16.70 |
| Energy contribution | -17.07 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.633057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
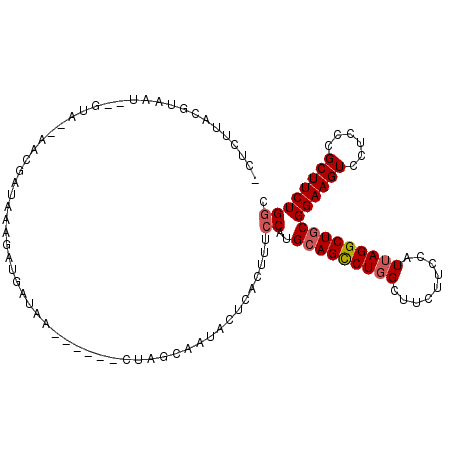
>3R_DroMel_CAF1 2191627 103 + 27905053 -CUCUUACGUAAAACCUAAUAACCAUAA--AUGAUAG------CCGGCAAUACUCACUUUCCGUGCAGCCUGGCUUCUUCCAUUAGGCUGCGGAAGUCCUCCCGCUUCUGGC -...........................--......(------((...................(((((((((.........)))))))))((((((......))))))))) ( -22.50) >DroPse_CAF1 14846 83 + 1 --------------------AUCGAUCA--AU--CAA---CCGCUA--AAUACUCACUUGCCAUGCAGCCUGGCCUCCUCCAUAAGGCUACGGAAGUCAUCGCGCUUCUGUC --------------------........--..--...---......--...........((.((((..((((((((........)))))).))..).))).))......... ( -14.50) >DroSec_CAF1 10887 105 + 1 -CUCUUACGUAAUACGUAAUAACUAUAAAGAUGAUAG------CCAGCAAUACUCACUUUCCAUGCAGCCUGGCUUCUUCCAUUAGGCUGCGGAAGUCCUCCCGCUUCUGGC -...(((((.....))))).................(------((((........((((((...(((((((((.........)))))))))))))))..........))))) ( -25.97) >DroSim_CAF1 10795 105 + 1 -CUCUUACGUAAUACGUAAUAACGAUAUAGAUGAUAG------CUAGCAAUACUCACUUUCCAUGCAGUCUGGCUUCUUCCAUUAGGCUGCGGAAGUCCUCCCGCUUCUGGC -...(((((.....))))).................(------((((........((((((...(((((((((.........)))))))))))))))..........))))) ( -20.97) >DroEre_CAF1 10796 102 + 1 ACCCUAACUUAAU--GUA--AACGAUAUAGAUGAUGA------UUAGCAAUACUCACUUUCCAUGCAGCCUGGCUUCUUCCAUUAGGCUGCGGAAGUCGUCACGCUUCUGGC .............--...--.........((((((((------.((....)).))...((((..(((((((((.........)))))))))))))))))))........... ( -23.40) >DroYak_CAF1 10252 102 + 1 -CCCUACCC---A--GUA--AA--AUAAAAAGGAGGAACAACGCUAGCAAUACUCACUUUCCAUGCAGCCUGGCUUCUUCCAUUAGGCUGCGGAAGUCCUCCCGCUUCUGGC -......((---(--(..--..--.......((((((.....(....).........(((((..(((((((((.........)))))))))))))))))))).....)))). ( -27.29) >consensus _CUCUUACGUAAU__GUA__AACGAUAAAGAUGAUAA______CUAGCAAUACUCACUUUCCAUGCAGCCUGGCUUCUUCCAUUAGGCUGCGGAAGUCCUCCCGCUUCUGGC ............................................................((..(((((((((.........)))))))))((((((......)))))))). (-16.70 = -17.07 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:51 2006