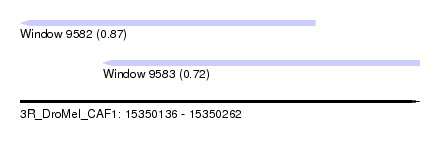
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,350,136 – 15,350,262 |
| Length | 126 |
| Max. P | 0.867646 |

| Location | 15,350,136 – 15,350,229 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 77.92 |
| Mean single sequence MFE | -18.18 |
| Consensus MFE | -11.74 |
| Energy contribution | -11.85 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.867646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
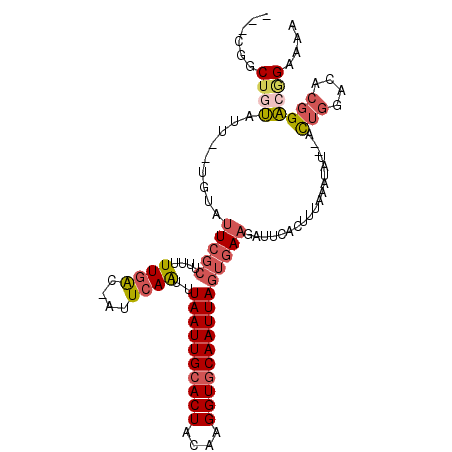
>3R_DroMel_CAF1 15350136 93 - 27905053 AUUUAAUUGCACUACAAGGUGCAAUUAGUGAAUAUUCACUUUAAAUAU--ACUGGACACGGACGGAAAAUGUAUUAUUAACACAUUGCAUUCAAA-------- ...((((((((((....))))))))))((((....)))).........--.(((....)))..(((.(((((.........)))))...)))...-------- ( -18.20) >DroVir_CAF1 126307 84 - 1 AAUUAAUUGCACUUCGCGGUGCAAUUAAUGAAAUUUG----------U--AUUCGACGCUGGUUGAAUGAAUGCAAGUGGC-------AUUUAAAUUUAUGGC .((((((((((((....)))))))))))).(((((((----------(--(((((((....))))))))(((((.....))-------))))))))))..... ( -25.20) >DroSec_CAF1 102976 93 - 1 AUUUAAUUGCACUACAAGGUGCAAUUAGUGAAGAGUCACUUUAAAUAU--ACUGGACACGGACGGAAAAUUUAUUAUUAACACAUAGCAUUCAAA-------- ...((((((((((....))))))))))((((....))))..(((((..--.(((........)))...)))))......................-------- ( -16.40) >DroSim_CAF1 106547 93 - 1 AUUUAAUUGCACUACAAGGUGCAAUUAGUGAAGAUUCACUUUAAAUAU--ACUGGACACGGACGGAAAAUUUAUUAUUAACACAUUGCAUUCAAA-------- ...((((((((((....))))))))))((((....))))..(((((..--.(((........)))...)))))......................-------- ( -16.40) >DroEre_CAF1 98938 93 - 1 GUUUAAUUGCACUACAAGGUGCAAUUAGUGAAGAUUAACUUUAAAAGU--ACUGGACACGGACGGAAAAUGUAUUAUUAACACAUUGCAUUCAAA-------- ..(((((((((((....)))))))))))(((((.....)))))...((--.(((....)))))(((.(((((.........)))))...)))...-------- ( -18.20) >DroYak_CAF1 129820 95 - 1 AUUUAAUUGCACUACAAGGUACAAUUAGUGAAGAUUCACUUUAAAUAUAGACUGGACACGGACAGAAAAUUUAUUAUUAACACCUUGCACUCAAA-------- ..............((((((.....(((((((..(((.((........)).(((....)))...)))..))))))).....))))))........-------- ( -14.70) >consensus AUUUAAUUGCACUACAAGGUGCAAUUAGUGAAGAUUCACUUUAAAUAU__ACUGGACACGGACGGAAAAUUUAUUAUUAACACAUUGCAUUCAAA________ ..(((((((((((....))))))))))).......................(((........)))...................................... (-11.74 = -11.85 + 0.11)


| Location | 15,350,162 – 15,350,262 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 79.87 |
| Mean single sequence MFE | -23.67 |
| Consensus MFE | -14.48 |
| Energy contribution | -15.52 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15350162 100 - 27905053 ---CGGCUGUAUU--UAUAUUCGCUUUUUUGAC-AUUCAAUUUAAUUGCACUACAAGGUGCAAUUAGUGAAUAUUCACUUUAAAUAU--ACUGGACACGGACGGAAAA ---...((((...--.((((((((....(((..-...)))..((((((((((....)))))))))))))))))).............--.(((....))))))).... ( -23.50) >DroVir_CAF1 126334 94 - 1 CAGCGCCUUCACUUUAGUCAU--CGUCUUUAGCUUGUCAAAUUAAUUGCACUUCGCGGUGCAAUUAAUGAAAUUUG----------U--AUUCGACGCUGGUUGAAUG ............((((..((.--((((....((..((...((((((((((((....))))))))))))...))..)----------)--....)))).))..)))).. ( -24.40) >DroSec_CAF1 103002 100 - 1 ---CGGCUGUAUU--UGUAUUCGCUUUUUUGAC-AUUCAAUUUAAUUGCACUACAAGGUGCAAUUAGUGAAGAGUCACUUUAAAUAU--ACUGGACACGGACGGAAAA ---((.((((.((--(((((.........((((-.((((...((((((((((....)))))))))).))))..))))........))--)).))).)))).))..... ( -25.33) >DroSim_CAF1 106573 100 - 1 ---CGGCUGUAUU--UGUGUUCGCUUUUUUGAC-AUUCAAUUUAAUUGCACUACAAGGUGCAAUUAGUGAAGAUUCACUUUAAAUAU--ACUGGACACGGACGGAAAA ---...((((...--((((((((....(((((.-........((((((((((....))))))))))((((....)))).)))))...--..)))))))).)))).... ( -26.50) >DroEre_CAF1 98964 100 - 1 ---UGGCUGUAUU--UGUAUUCGCUUUUUUGAC-AUUCAGUUUAAUUGCACUACAAGGUGCAAUUAGUGAAGAUUAACUUUAAAAGU--ACUGGACACGGACGGAAAA ---...((((...--(((....((((((((((.-.((((..(((((((((((....)))))))))))))))..))))....))))))--.....)))...)))).... ( -23.60) >DroYak_CAF1 129846 102 - 1 ---UGGCUUUGUU--UGUAUUCGCUUUUUUGAC-AUUCAAUUUAAUUGCACUACAAGGUACAAUUAGUGAAGAUUCACUUUAAAUAUAGACUGGACACGGACAGAAAA ---....((((((--(((.(((.((..(((((.-........((((((.(((....))).))))))((((....)))).)))))...))...))).)))))))))... ( -18.70) >consensus ___CGGCUGUAUU__UGUAUUCGCUUUUUUGAC_AUUCAAUUUAAUUGCACUACAAGGUGCAAUUAGUGAAGAUUCACUUUAAAUAU__ACUGGACACGGACGGAAAA ......((((.........(((((....((((....))))..((((((((((....)))))))))))))))...................(((....))))))).... (-14.48 = -15.52 + 1.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:22 2006