
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,330,511 – 15,330,776 |
| Length | 265 |
| Max. P | 0.982594 |

| Location | 15,330,511 – 15,330,607 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 80.52 |
| Mean single sequence MFE | -36.30 |
| Consensus MFE | -23.78 |
| Energy contribution | -23.67 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15330511 96 + 27905053 GGACGAGCAGGUGACUCGCGACAAUCCCUGGAGGAACUGAAACAACUGAGGGCCCAGGCCCAGCAGCAAUCCCUGCUCAAUAAUUCCUCCUCAUCA------ (((...((((....)).)).....)))..(((((((.............((((....))))(((((......)))))......)))))))......------ ( -32.60) >DroVir_CAF1 102150 102 + 1 AGUCGCGCCGGCGACUCGCGGCAAUCCCUGGAGGAACUGAAGCAGCUGCGUGCCCAGGCGCAGCAGCAGUCCCUGCACAAUUCAUCCUCCUCGUCCGUCUCA ((((((....)))))).((((........((((((..((((...(((((((......))))))).((((...))))....))))))))))....)))).... ( -39.80) >DroPse_CAF1 110294 96 + 1 GGACGUGCCGGAGACUCGCGACAAUCAUUGGAGGAGCUGAAGCAACUGCGGGCCCAGGCCCAACAGCAAUCGCUGCACAACUCCUCCUCCUCUUCG------ (((((((..(....).)))).........(((((((.((..((....))((((....))))..((((....))))..)).))))))))))......------ ( -37.50) >DroMoj_CAF1 97958 102 + 1 AGCCGUGCAGGAGACUCGCGUCAGUCGCUGGAGGAACUGAAGCAGCUGCGUGCCCAGGCGCAGCAGCAAUCGCUACACAAUUCAUCCUCCUCGUCUGUGUCA ...((((.((....)))))).(((.((..((((((..((((...(((((((......)))))))(((....)))......)))))))))).)).)))..... ( -39.90) >DroAna_CAF1 77119 93 + 1 GGCCGAGCGGGGGACUCGCGACAAUCACUGGAGGAACUGAAGCAACUGAGGGCCCAGGCCCAGCAACAAUCCAUGCACA---ACUCCUCCUCCUCG------ ...(((((((....).)))..........((((((..((..........((((....)))).(((........))).))---..))))))...)))------ ( -30.50) >DroPer_CAF1 106220 96 + 1 GGACGUGCCGGAGACUCGCGACAAUCAUUGGAGGAGCUGAAGCAACUGCGGGCCCAGGCCCAACAGCAAUCGCUGCACAACUCCUCCUCCUCUUCG------ (((((((..(....).)))).........(((((((.((..((....))((((....))))..((((....))))..)).))))))))))......------ ( -37.50) >consensus GGACGUGCCGGAGACUCGCGACAAUCACUGGAGGAACUGAAGCAACUGCGGGCCCAGGCCCAGCAGCAAUCCCUGCACAAUUCCUCCUCCUCGUCG______ .........(((((.....(.....)...((((((..............((((....)))).((((......))))........)))))))))))....... (-23.78 = -23.67 + -0.11)



| Location | 15,330,551 – 15,330,644 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 76.85 |
| Mean single sequence MFE | -28.15 |
| Consensus MFE | -17.89 |
| Energy contribution | -19.62 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823448 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15330551 93 + 27905053 AACAACUGAGGGCCCAGGCCCAGCAGCAAUCCCUGCUCAAUAAUUCCUCCUCAUCA---------AGCUCCAAUUCCCAGGUGGAGCAAUCCAUGCGCUCCA .....(((.((((....))))(((((......)))))...................---------............))).((((((.........)))))) ( -25.00) >DroVir_CAF1 102190 102 + 1 AGCAGCUGCGUGCCCAGGCGCAGCAGCAGUCCCUGCACAAUUCAUCCUCCUCGUCCGUCUCAUCCGCUUCGGCCUCGCAGGUGGAGCAGUCCAUGCGCUCCA ....(((((((((....)))).)))))....(((((................(.......)....((....))...)))))((((((.((....)))))))) ( -35.10) >DroPse_CAF1 110334 90 + 1 AGCAACUGCGGGCCCAGGCCCAACAGCAAUCGCUGCACAACUCCUCCUCCUCUUCG---------UCAUC---CUCGCACGUGGAACAGUCCAUGCGGUCCA ....(((((((((....))))..((((....)))).....................---------.....---.......(((((....))))))))))... ( -23.80) >DroMoj_CAF1 97998 102 + 1 AGCAGCUGCGUGCCCAGGCGCAGCAGCAAUCGCUACACAAUUCAUCCUCCUCGUCUGUGUCAUCCGCAUCGGCCUCGCAGGUGGAGCAAUCAAUGCGCUCCA .((((((((((((....)))).)))))...................((((...((((((....(((...)))...)))))).)))).......)))...... ( -32.90) >DroAna_CAF1 77159 90 + 1 AGCAACUGAGGGCCCAGGCCCAGCAACAAUCCAUGCACA---ACUCCUCCUCCUCG---------AGCUCCAACUCCCAGGUGGAGCAGUCCAUGCGAUCCA .(((((((.((((....)))).(((........)))...---....((((.(((.(---------((......)))..))).))))))))...)))...... ( -28.30) >DroPer_CAF1 106260 90 + 1 AGCAACUGCGGGCCCAGGCCCAACAGCAAUCGCUGCACAACUCCUCCUCCUCUUCG---------UCAUC---CUCGCACGUGGAACAGUCCAUGCGGUCCA ....(((((((((....))))..((((....)))).....................---------.....---.......(((((....))))))))))... ( -23.80) >consensus AGCAACUGCGGGCCCAGGCCCAGCAGCAAUCCCUGCACAAUUCCUCCUCCUCGUCG_________ACAUC___CUCGCAGGUGGAGCAGUCCAUGCGCUCCA .((..((((((((....)))).((((......))))........................................))))(((((....)))))))...... (-17.89 = -19.62 + 1.72)

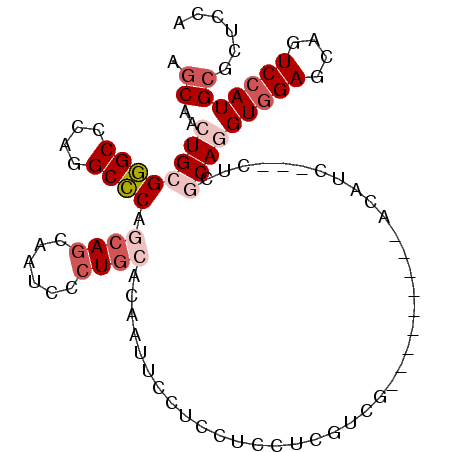

| Location | 15,330,585 – 15,330,684 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 73.71 |
| Mean single sequence MFE | -25.90 |
| Consensus MFE | -16.61 |
| Energy contribution | -17.75 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15330585 99 + 27905053 GCUCAAUAAUUCCUCCUCAUCA------------AGCUCCAAUUCCCAGGUGGAGCAAUCCAUGCGCUCCACCAUUGAGCACCAUAUGCGCUCACUGGAUCGCAACUUACC (((((((........((.....------------))............((((((((.........)))))))))))))))......((((.((....)).))))....... ( -28.40) >DroVir_CAF1 102224 108 + 1 GCACAAUUCAUCCUCCUCGUCC---GUCUCAUCCGCUUCGGCCUCGCAGGUGGAGCAGUCCAUGCGCUCCACCAUCGAACAUCACAUGCGCUCGCUGGAUCGCAAUCUGCC (((......((((....((..(---((.......((....)).(((..((((((((.((....))))))))))..))).........)))..))..)))).......))). ( -29.82) >DroPse_CAF1 110368 96 + 1 GCACAACUCCUCCUCCUCUUCG------------UCAUC---CUCGCACGUGGAACAGUCCAUGCGGUCCACCAUUGAGCAUCACAUGAGAUCCCUGGAUCGCAAUCUUCC ((...................(------------(.((.---(((...((((((....)))))).((....))...))).)).))....((((....))))))........ ( -19.50) >DroWil_CAF1 111855 105 + 1 GCAGCACAAUUCCUCAUCAUCGUCAGUG------UCAUCCAAUUCACAUGUCGAGCAAUCGAUGCGCUCCACCAUUGAGCAUCAUAUGCGUUCACUUGAUCGCAAUUUACC ...((((.(((.(((..(((.((.(((.------.......))).)))))..))).))).).)))((((.......))))......((((.((....)).))))....... ( -20.80) >DroMoj_CAF1 98032 108 + 1 ACACAAUUCAUCCUCCUCGUCU---GUGUCAUCCGCAUCGGCCUCGCAGGUGGAGCAAUCAAUGCGCUCCACCAUCGAGCACCACAUGCGCUCCCUCGAUCGCAAUCUGCC (((((................)---)))).....(((..((.((((..((((((((.........))))))))..))))..))...((((.((....)).))))...))). ( -31.09) >DroAna_CAF1 77193 96 + 1 GCACA---ACUCCUCCUCCUCG------------AGCUCCAACUCCCAGGUGGAGCAGUCCAUGCGAUCCACCAUCGAGCACCACAUGCGAUCCCUGGACCGCAACCUACC .....---(((.((((.(((.(------------((......)))..))).)))).)))...((((.((((.......(((.....)))......)))).))))....... ( -25.82) >consensus GCACAAUUAAUCCUCCUCAUCG____________ACAUCCAACUCGCAGGUGGAGCAAUCCAUGCGCUCCACCAUCGAGCACCACAUGCGCUCCCUGGAUCGCAAUCUACC ..........................................((((..((((((((.........))))))))..)))).......((((.((....)).))))....... (-16.61 = -17.75 + 1.14)



| Location | 15,330,607 – 15,330,724 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.90 |
| Mean single sequence MFE | -36.45 |
| Consensus MFE | -23.00 |
| Energy contribution | -23.03 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15330607 117 + 27905053 ---AGCUCCAAUUCCCAGGUGGAGCAAUCCAUGCGCUCCACCAUUGAGCACCAUAUGCGCUCACUGGAUCGCAACUUACCACUGGAGCUCCAAUAUAGUCGCCAUCGCUUUCAGAACAAU ---(((((((........(((((....)))))(((.((((....(((((.(.....).))))).)))).)))..........)))))))...........((....))............ ( -32.90) >DroVir_CAF1 102252 120 + 1 UCCGCUUCGGCCUCGCAGGUGGAGCAGUCCAUGCGCUCCACCAUCGAACAUCACAUGCGCUCGCUGGAUCGCAAUCUGCCGCUGGAGCUGCAAUACAGCCGCCAUCGCAUGCAGAACAAU ...((((((((..((((((((((((.((....))))))))))...(.....)...))))...))))))..))..(((((((.(((.((((.....))))..))).))...)))))..... ( -43.70) >DroGri_CAF1 102156 120 + 1 UCCGCCUCCGCCUCGCAGGUGGAGCAAUCGAUGCGCUCCACUAUUGAGCAUCAUAUGCGUUCGCUGGACCGCAAUCUGCCGCUGGAGCUGCAGUAUAGUAGGCAUCGCAUGCAAAACAAU (((((((.((...)).)))))))(((..((((((((((.......))))......((((.((....)).))))..((((.((....)).))))........))))))..)))........ ( -41.90) >DroWil_CAF1 111883 117 + 1 ---UCAUCCAAUUCACAUGUCGAGCAAUCGAUGCGCUCCACCAUUGAGCAUCAUAUGCGUUCACUUGAUCGCAAUUUACCACUGGAAUUGCAAUAUAGUCGUCAUCGUUUUCAAAAUAAC ---.....((((((.((.(((((....)))))..((((.......))))......((((.((....)).)))).........)))))))).............................. ( -21.80) >DroMoj_CAF1 98060 120 + 1 UCCGCAUCGGCCUCGCAGGUGGAGCAAUCAAUGCGCUCCACCAUCGAGCACCACAUGCGCUCCCUCGAUCGCAAUCUGCCGCUGGAACUGCAGUACAGUCGCCAUCGCAUGCAGAACAAC ((.((((.(((((((..((((((((.........))))))))..)))).......((((.((....)).))))....)))(((((.((((.....))))..)))..)))))).))..... ( -40.90) >DroAna_CAF1 77212 117 + 1 ---AGCUCCAACUCCCAGGUGGAGCAGUCCAUGCGAUCCACCAUCGAGCACCACAUGCGAUCCCUGGACCGCAACCUACCCCUGGAGCUGCAGUACAGCCGUCAUCGGUUCCAGAACAAC ---(((((((.(((...(((((((((.....)))..))))))...))).......((((.((....)).)))).........))))))).......(((((....))))).......... ( -37.50) >consensus ___GCCUCCAACUCGCAGGUGGAGCAAUCCAUGCGCUCCACCAUCGAGCACCACAUGCGCUCACUGGAUCGCAAUCUACCGCUGGAGCUGCAAUACAGUCGCCAUCGCAUGCAGAACAAC ......((((.((((..((((((((.........))))))))..)))).......((((.((....)).)))).........))))((((.....))))..................... (-23.00 = -23.03 + 0.03)

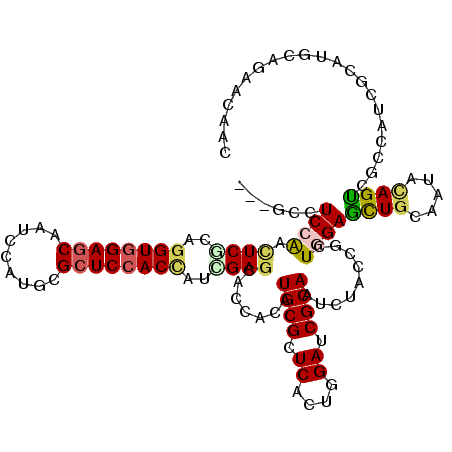

| Location | 15,330,607 – 15,330,724 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.90 |
| Mean single sequence MFE | -44.97 |
| Consensus MFE | -26.22 |
| Energy contribution | -25.08 |
| Covariance contribution | -1.14 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.58 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15330607 117 - 27905053 AUUGUUCUGAAAGCGAUGGCGACUAUAUUGGAGCUCCAGUGGUAAGUUGCGAUCCAGUGAGCGCAUAUGGUGCUCAAUGGUGGAGCGCAUGGAUUGCUCCACCUGGGAAUUGGAGCU--- ...((((..(..(.(((.((((((.((((((....))))))...)))))).))))..(((((((.....)))))))..(((((((((.......)))))))))......)..)))).--- ( -49.90) >DroVir_CAF1 102252 120 - 1 AUUGUUCUGCAUGCGAUGGCGGCUGUAUUGCAGCUCCAGCGGCAGAUUGCGAUCCAGCGAGCGCAUGUGAUGUUCGAUGGUGGAGCGCAUGGACUGCUCCACCUGCGAGGCCGAAGCGGA .....(((((......(((.(((((.....)))))))).((((...(((((......(((((((....).))))))..(((((((((.......)))))))))))))).))))..))))) ( -50.00) >DroGri_CAF1 102156 120 - 1 AUUGUUUUGCAUGCGAUGCCUACUAUACUGCAGCUCCAGCGGCAGAUUGCGGUCCAGCGAACGCAUAUGAUGCUCAAUAGUGGAGCGCAUCGAUUGCUCCACCUGCGAGGCGGAGGCGGA ........(((..(((((((((((((.((((.((....)).)))).((((......))))..((((...))))...)))))))...))))))..)))(((.(((.((...)).))).))) ( -41.20) >DroWil_CAF1 111883 117 - 1 GUUAUUUUGAAAACGAUGACGACUAUAUUGCAAUUCCAGUGGUAAAUUGCGAUCAAGUGAACGCAUAUGAUGCUCAAUGGUGGAGCGCAUCGAUUGCUCGACAUGUGAAUUGGAUGA--- ((((((.(....).))))))..............((((((.(((...((((.((....)).)))).....)))(((...((.(((((.......))))).))...)))))))))...--- ( -27.50) >DroMoj_CAF1 98060 120 - 1 GUUGUUCUGCAUGCGAUGGCGACUGUACUGCAGUUCCAGCGGCAGAUUGCGAUCGAGGGAGCGCAUGUGGUGCUCGAUGGUGGAGCGCAUUGAUUGCUCCACCUGCGAGGCCGAUGCGGA .....(((((((....(((.(((((.....)))))))).((((...(((((.......((((((.....))))))...(((((((((.......)))))))))))))).))))))))))) ( -53.70) >DroAna_CAF1 77212 117 - 1 GUUGUUCUGGAACCGAUGACGGCUGUACUGCAGCUCCAGGGGUAGGUUGCGGUCCAGGGAUCGCAUGUGGUGCUCGAUGGUGGAUCGCAUGGACUGCUCCACCUGGGAGUUGGAGCU--- ((((((.((....))..))))))....((.(((((((..((((.((..(((((((((.((((.(((((.(....).)).))))))).).)))))))).)))))).))))))).))..--- ( -47.50) >consensus AUUGUUCUGCAAGCGAUGGCGACUAUACUGCAGCUCCAGCGGCAGAUUGCGAUCCAGCGAGCGCAUAUGAUGCUCAAUGGUGGAGCGCAUGGAUUGCUCCACCUGCGAGGCGGAGGC___ .....(((((..((((...........((((.((....)).))))..((((.((....)).))))........))...(((((((((.......))))))))).))...)))))...... (-26.22 = -25.08 + -1.14)

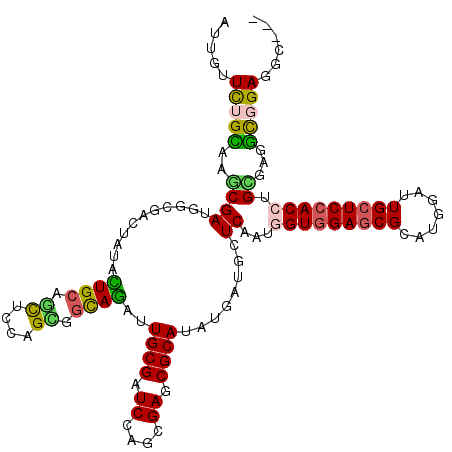
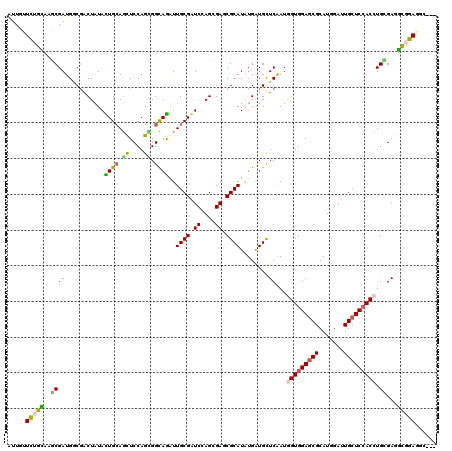
| Location | 15,330,684 – 15,330,776 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 78.19 |
| Mean single sequence MFE | -37.42 |
| Consensus MFE | -20.70 |
| Energy contribution | -21.40 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15330684 92 + 27905053 ACUGGAGCUCCAAUAUAGUCGCCAUCGCUUUCAGAACAAUUUGAAUGCCGUGGCCGCUGCUG------GUGGUGGUGG---U------------GGAAAUUCGUCC---ACCGGAA ......((((((...((((.(((((.((.((((((....)))))).)).))))).))))...------.))).)))((---(------------(((......)))---))).... ( -33.00) >DroSec_CAF1 83505 92 + 1 CCUGGAGCUGCAAUAUAGUCGCCAUCGCUUUCAGAACAAUUUGAAUGCCGUGGCCGCUGCUG------CAGGUGGUGC---C------------GCAAACUCGUCC---ACCGGAA .((((..(((((...((((.(((((.((.((((((....)))))).)).))))).)))).))------)))(((....---)------------))..........---.)))).. ( -33.10) >DroSim_CAF1 85284 92 + 1 ACUGGAGCUCCAAUAUAGUCGCCAUCGCUUUCAGAACAAUUUGAAUGCCGUGGCCGCUGCUG------CAGGUGGUGC---C------------GCAAACUCGUCC---ACCGGAA ........(((....((((.(((((.((.((((((....)))))).)).))))).))))...------..(((((...---(------------(......)).))---)))))). ( -30.00) >DroEre_CAF1 79110 101 + 1 ACUGGAGCUGCAGUAUAGUCGCCAUCGCUUUCAGAACAAUUUGAAUGCCGUGGCCGCUGCUGCGGCUGCUGGCAGUGCUGGU------------GGCAACUCGUCC---ACCGGAA .(..((((((((((..(((.(((((.((.((((((....)))))).)).))))).)))))))))))).)..).....(((((------------(((.....).))---))))).. ( -47.10) >DroYak_CAF1 90401 113 + 1 ACUGGAGCUGCAAUAUAGUCGCCAUCGCUUUCAGAACAAUUUGAAUGCCGUGGCCGCUGCCGCUGCUGCAGGUGGUGCUGGUGGUGGUGGUGUUGGCAAUUCGUCC---ACCGGAA .(((.(((.((....((((.(((((.((.((((((....)))))).)).))))).))))..)).))).)))......((((((((((...((....))..))).))---))))).. ( -43.80) >DroAna_CAF1 77289 95 + 1 CCUGGAGCUGCAGUACAGCCGUCAUCGGUUCCAGAACAACCUGAAUGCCGUGGCCGCUGCAU------CUGCUGCGAG---C------------GGGAAUCCUUCCGCAACUGUCA ......((.((((..((((.((((.((((..(((......)))...)))))))).))))...------)))).))..(---(------------((((....))))))........ ( -37.50) >consensus ACUGGAGCUGCAAUAUAGUCGCCAUCGCUUUCAGAACAAUUUGAAUGCCGUGGCCGCUGCUG______CAGGUGGUGC___C____________GGAAACUCGUCC___ACCGGAA .((((...............(((((.((.((((((....)))))).)).)))))(((((((.........))))))).................................)))).. (-20.70 = -21.40 + 0.70)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:06:14 2006