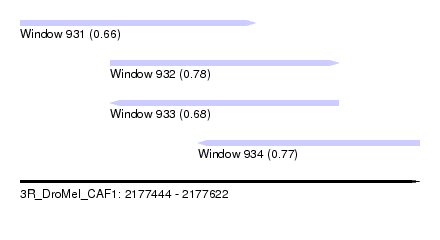
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,177,444 – 2,177,622 |
| Length | 178 |
| Max. P | 0.783766 |

| Location | 2,177,444 – 2,177,549 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 84.40 |
| Mean single sequence MFE | -23.24 |
| Consensus MFE | -15.88 |
| Energy contribution | -16.28 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2177444 105 + 27905053 AGACAUUUACAUAAAGCUCUAAUAACUAACAACAUUUCGCAGUCGAUUUGGCAUUUAACAGUUACCGCUCUAACGGCACAUGUCCAGUGCGCC-AGUGC-------ACCACCC------ ...............((.((............((..(((....)))..))(((((..(((....(((......)))....)))..)))))...-)).))-------.......------ ( -17.30) >DroSec_CAF1 2536 107 + 1 AGACAUUUACAUAAAGCUCUAAUAACUAACAACAUUUCGCAGUCGAUUUGGCAUUUAACAGUUGCCGCUCUAACGGCACAUGUCCAGUGCCCCUAUCGC------UACCACCC------ ........................................((.((((..((((((..(((..(((((......)))))..)))..))))))...)))))------).......------ ( -23.30) >DroSim_CAF1 2645 107 + 1 AGACAUUUACAUAAAGCUCUAAUAACUAACAACAUUUCGCAGUCGAUUUGGCAUUUAACAGUUGCCGCUCUAACGGCACAUGUCCAGUGCCCCUAGCGC------UACCACCU------ ..............(((.(((...............(((....)))...((((((..(((..(((((......)))))..)))..))))))..))).))------).......------ ( -23.50) >DroEre_CAF1 4323 113 + 1 AGACAUUUACAUAAAGCUC-AAUAACUAACAACAUUUCGCACUCGAUUUGGCAUUUAACAGUUGCCGCUAUAACGGCACAUGUCCAGUGCCCCUAGCUCCCCC-----CUCCGGGGGGC ..............((((.-................(((....)))...((((((..(((..(((((......)))))..)))..))))))...))))(((((-----(...)))))). ( -32.30) >DroYak_CAF1 2854 106 + 1 AGACAUUUACAUAAAGUUC-CAUAGCUAAUAACAUUU------CGAUUUGGCAUUUAACAGUUGCCGCUAUAACGGCACAUGUCCAGUGCCCCAUGCUCAUCCUUUUCCUCCC------ .(((((........(((..-....)))..........------......((((((..(((..(((((......)))))..)))..))))))..))).))..............------ ( -19.80) >consensus AGACAUUUACAUAAAGCUCUAAUAACUAACAACAUUUCGCAGUCGAUUUGGCAUUUAACAGUUGCCGCUCUAACGGCACAUGUCCAGUGCCCCUAGCGC______UACCACCC______ .................................................((((((..(((..(((((......)))))..)))..))))))............................ (-15.88 = -16.28 + 0.40)


| Location | 2,177,484 – 2,177,586 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.02 |
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -15.88 |
| Energy contribution | -16.28 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.783766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2177484 102 + 27905053 AGUCGAUUUGGCAUUUAACAGUUACCGCUCUAACGGCACAUGUCCAGUGCGCC-AGUGC-------ACCACCC---------CCUUCCUUCGUUCAACGAAAAACAUAUUUGCUUAGCA (((.(((.(((((((.....((((......))))(((.(((.....))).)))-)))))-------.......---------......((((.....))))...)).))).)))..... ( -18.50) >DroSec_CAF1 2576 104 + 1 AGUCGAUUUGGCAUUUAACAGUUGCCGCUCUAACGGCACAUGUCCAGUGCCCCUAUCGC------UACCACCC---------CCUUCCCUCGUUCGAGGAAAACCAUAUUUACUUAGCA ((.((((..((((((..(((..(((((......)))))..)))..))))))...)))))------).......---------.....((((....)))).................... ( -28.60) >DroSim_CAF1 2685 104 + 1 AGUCGAUUUGGCAUUUAACAGUUGCCGCUCUAACGGCACAUGUCCAGUGCCCCUAGCGC------UACCACCU---------CCUUCCCUCGUUCGAGGAAAACCAUAUUUGCUUAGCA .........((((((..(((..(((((......)))))..)))..))))))......((------((......---------.....((((....)))).....((....))..)))). ( -26.60) >DroEre_CAF1 4362 114 + 1 ACUCGAUUUGGCAUUUAACAGUUGCCGCUAUAACGGCACAUGUCCAGUGCCCCUAGCUCCCCC-----CUCCGGGGGGCAGGGCCUCCUCCUUUCGUGGAAAAACUUAUUUGCUUAGCA .........((((((..(((..(((((......)))))..)))..))))))(((.(((((((.-----....))))))))))......(((......)))................... ( -38.50) >DroYak_CAF1 2890 107 + 1 ---CGAUUUGGCAUUUAACAGUUGCCGCUAUAACGGCACAUGUCCAGUGCCCCAUGCUCAUCCUUUUCCUCCC---------UCUUCCUGCUUUCGAGCAAAAACAUAUUUGUUUAGCU ---......((((((..(((..(((((......)))))..)))..))))))....(((...............---------......((((....)))).(((((....)))))))). ( -25.20) >consensus AGUCGAUUUGGCAUUUAACAGUUGCCGCUCUAACGGCACAUGUCCAGUGCCCCUAGCGC______UACCACCC_________CCUUCCUUCGUUCGAGGAAAAACAUAUUUGCUUAGCA .........((((((..(((..(((((......)))))..)))..)))))).................................................................... (-15.88 = -16.28 + 0.40)

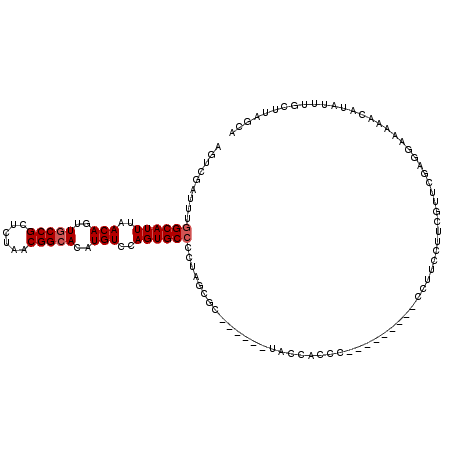
| Location | 2,177,484 – 2,177,586 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.02 |
| Mean single sequence MFE | -34.16 |
| Consensus MFE | -20.72 |
| Energy contribution | -20.88 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2177484 102 - 27905053 UGCUAAGCAAAUAUGUUUUUCGUUGAACGAAGGAAGG---------GGGUGGU-------GCACU-GGCGCACUGGACAUGUGCCGUUAGAGCGGUAACUGUUAAAUGCCAAAUCGACU .((((.(((..(((.((((((.((.....)).)))))---------).))).)-------))..)-)))(((...((((..((((((....))))))..))))...))).......... ( -26.60) >DroSec_CAF1 2576 104 - 1 UGCUAAGUAAAUAUGGUUUUCCUCGAACGAGGGAAGG---------GGGUGGUA------GCGAUAGGGGCACUGGACAUGUGCCGUUAGAGCGGCAACUGUUAAAUGCCAAAUCGACU .((((.....((....((((((((....)))))))).---------..))..))------))(((...((((...((((..((((((....))))))..))))...))))..))).... ( -34.70) >DroSim_CAF1 2685 104 - 1 UGCUAAGCAAAUAUGGUUUUCCUCGAACGAGGGAAGG---------AGGUGGUA------GCGCUAGGGGCACUGGACAUGUGCCGUUAGAGCGGCAACUGUUAAAUGCCAAAUCGACU (((...))).......((((((((....))))))))(---------(..(((((------((.((((.(((((.......))))).)))).))(....).......)))))..)).... ( -36.10) >DroEre_CAF1 4362 114 - 1 UGCUAAGCAAAUAAGUUUUUCCACGAAAGGAGGAGGCCCUGCCCCCCGGAG-----GGGGGAGCUAGGGGCACUGGACAUGUGCCGUUAUAGCGGCAACUGUUAAAUGCCAAAUCGAGU .(((((((......)))).(((.(....)..))))))((((((((((....-----))))).)).)))((((...((((..((((((....))))))..))))...))))......... ( -44.80) >DroYak_CAF1 2890 107 - 1 AGCUAAACAAAUAUGUUUUUGCUCGAAAGCAGGAAGA---------GGGAGGAAAAGGAUGAGCAUGGGGCACUGGACAUGUGCCGUUAUAGCGGCAACUGUUAAAUGCCAAAUCG--- .(((...........(((((.(((...........))---------).)))))........)))....((((...((((..((((((....))))))..))))...))))......--- ( -28.61) >consensus UGCUAAGCAAAUAUGUUUUUCCUCGAACGAAGGAAGG_________GGGUGGUA______GCGCUAGGGGCACUGGACAUGUGCCGUUAGAGCGGCAACUGUUAAAUGCCAAAUCGACU .................(((((((....).))))))................................((((...((((..((((((....))))))..))))...))))......... (-20.72 = -20.88 + 0.16)

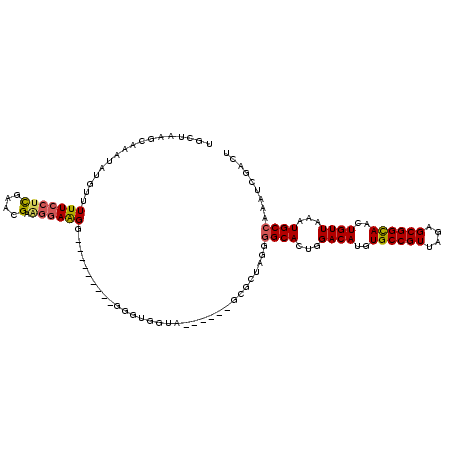
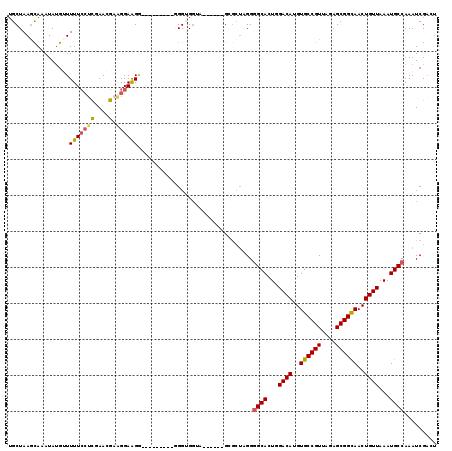
| Location | 2,177,523 – 2,177,622 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.67 |
| Mean single sequence MFE | -40.50 |
| Consensus MFE | -17.96 |
| Energy contribution | -18.72 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2177523 99 - 27905053 ----GCGCGUCCCUGUCCCUUCCGCUUUUACCUGUGACCGUGCUAAGCAAAUAUGUUUUUCGUUGAACGAAGGAAGG---------GGGUGGU-------GCACU-GGCGCACUGGACAU ----....((((...((((((((((((.(((........)))..))))..........((((.....))))))))))---------))..(((-------((...-...))))))))).. ( -33.60) >DroSec_CAF1 2615 101 - 1 ----GCGCGUCCCUGUCCCUUCCGCUUUUACCUGUGACCGUGCUAAGUAAAUAUGGUUUUCCUCGAACGAGGGAAGG---------GGGUGGUA------GCGAUAGGGGCACUGGACAU ----....((((.(((((((..((((.((((((.((((((((.........)))))))((((((....)))))).).---------)))))).)------)))..)))))))..)))).. ( -43.90) >DroSim_CAF1 2724 101 - 1 ----GCGCGUCCCUGUCCCUUCCGCUUUUACCUGUGACCGUGCUAAGCAAAUAUGGUUUUCCUCGAACGAGGGAAGG---------AGGUGGUA------GCGCUAGGGGCACUGGACAU ----....((((.(((((((..((((.((((((.((((((((.........)))))))((((((....)))))).).---------)))))).)------)))..)))))))..)))).. ( -43.90) >DroEre_CAF1 4401 115 - 1 CUCCCCGCGUCCCUGUCCCAUCCGCUUUUACCUGUGACCGUGCUAAGCAAAUAAGUUUUUCCACGAAAGGAGGAGGCCCUGCCCCCCGGAG-----GGGGGAGCUAGGGGCACUGGACAU ........((((.((((((....(((((..(((.....((((..((((......))))...))))..)))..)))))...(((((((....-----))))).))..))))))..)))).. ( -45.00) >DroYak_CAF1 2926 107 - 1 ----ACGCGUCCCUCUCCCAACCGCUUUUACCUGUGACCGAGCUAAACAAAUAUGUUUUUGCUCGAAAGCAGGAAGA---------GGGAGGAAAAGGAUGAGCAUGGGGCACUGGACAU ----....(((((.(((((..((.(((((.(((((...(((((.(((((....)))))..)))))...))))).)))---------))..))....))..)))...)))))......... ( -36.10) >consensus ____GCGCGUCCCUGUCCCUUCCGCUUUUACCUGUGACCGUGCUAAGCAAAUAUGUUUUUCCUCGAACGAAGGAAGG_________GGGUGGUA______GCGCUAGGGGCACUGGACAU ........((((.((((((...(((........))).((.(((...)))........(((((((....).))))))...........)).................))))))..)))).. (-17.96 = -18.72 + 0.76)

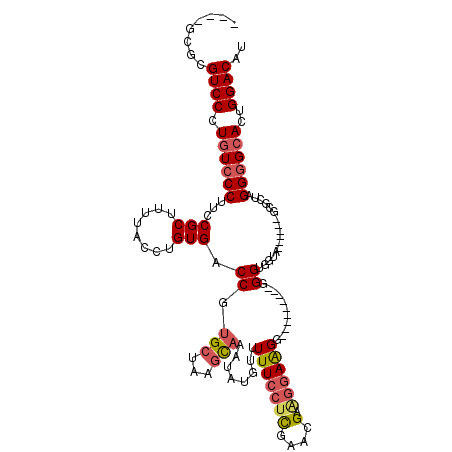
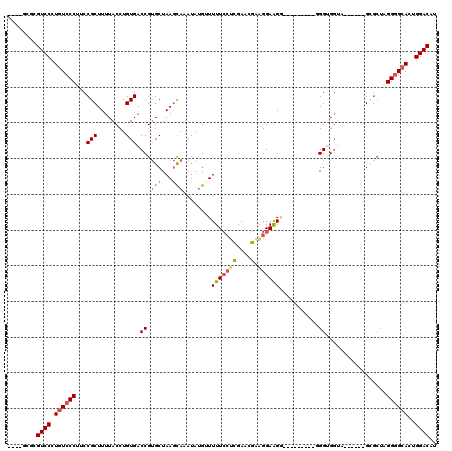
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:45 2006