
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,287,854 – 15,288,014 |
| Length | 160 |
| Max. P | 0.788409 |

| Location | 15,287,854 – 15,287,974 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.67 |
| Mean single sequence MFE | -46.76 |
| Consensus MFE | -41.60 |
| Energy contribution | -41.28 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752823 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
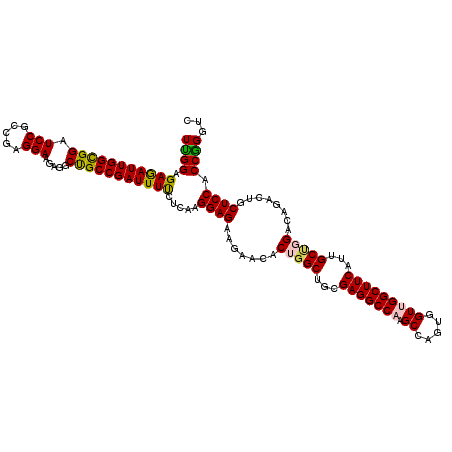
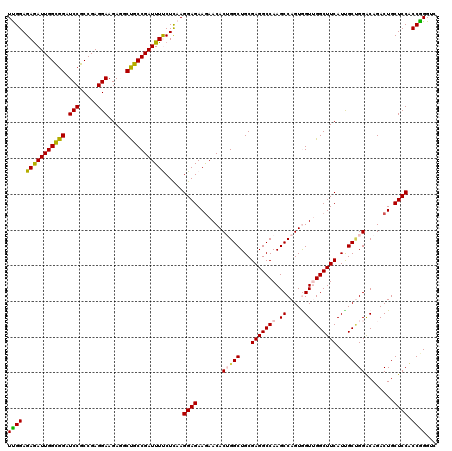
>3R_DroMel_CAF1 15287854 120 - 27905053 UUGGAGAGAUUGGCGGAUCCGCCGAGGAAGAGGCUGCCGAUUUCCUUAAGGAGAAGAACACUGGCUGCGAGGCCAAGCCAGUAGUUGGCUUCAUUGCUGGACAGACUGCUCCACCGGGUC ((((.(((((((((((.(((.....))).....))))))))))).....((((.((....(..((...((((((((........))))))))...))..).....)).)))).))))... ( -44.00) >DroSec_CAF1 40397 120 - 1 UCGGAGAGAUUGGCGGAUCCGCCGAGGAAGAUGCUGCCGAUUUUCUCAAGGAGAAGAACACUGGCUGCGAGGCCAAGCCAGUGGUGGGCUUCAUUGCUGGACAGACUGCUCCACCGGGUC ((((.((((((((((((((..(....)..))).)))))))...))))..((((.((.....(((((....))))).(((((..(((.....)))..)))).)...)).)))).))))... ( -46.90) >DroSim_CAF1 41089 120 - 1 UUGGAGAAAUUGGCGGAUCCGCCGAGGAAGAUGCUGCCGAUUUUCUCAAGGAGAAGAACACUGGCUGCGAGGCCAAGCCAGUGGUGGGCUUCAUUGCUGGGCAGACUGCUCCACCGGGUC .(((((...((((((....))))))........(((((..(((((((...))))))).((((((((.........))))))))...(((......))).)))))....)))))....... ( -46.80) >DroEre_CAF1 36489 120 - 1 UCGGAGAGAUUGGCGGAUCCGCCGAGGAAGAGGCCGCCGAUUUUCUCAAGGAGAAAAACACAGGCUGCGAGGCCAAGCCAGUAGUUGGCUUCAUCGCAGGACAGACUGCUCCUCCGGGUC ((((((.((((((((....)))))).((.((((((...(.(((((((...))))))).)...((((((..(((...))).)))))))))))).))((((......))))))))))))... ( -50.70) >DroYak_CAF1 39659 120 - 1 UUGGAGAGAUUGGUGGAUCCGCCGAGGAAGAAGCCGCCGAUUUCCUCAAGGAGAAGAACACAGGCUGCGAGGCCAAGCCAGUGGUUGGCUUCAUCGCCGGACAGACUGCUCCCCCAGGUC (((..(((((((((((.(((.....))).....)))))))))))..)))((((.((......(((...((((((((.(....).))))))))...))).......)).))))........ ( -45.42) >consensus UUGGAGAGAUUGGCGGAUCCGCCGAGGAAGAGGCUGCCGAUUUUCUCAAGGAGAAGAACACUGGCUGCGAGGCCAAGCCAGUGGUUGGCUUCAUUGCUGGACAGACUGCUCCACCGGGUC ((((.(((((((((((.(((.....))).....))))))))))).....((((.......(((((...(((((((.((.....)))))))))...)))))........)))).))))... (-41.60 = -41.28 + -0.32)

| Location | 15,287,894 – 15,288,014 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -40.01 |
| Consensus MFE | -36.00 |
| Energy contribution | -35.28 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
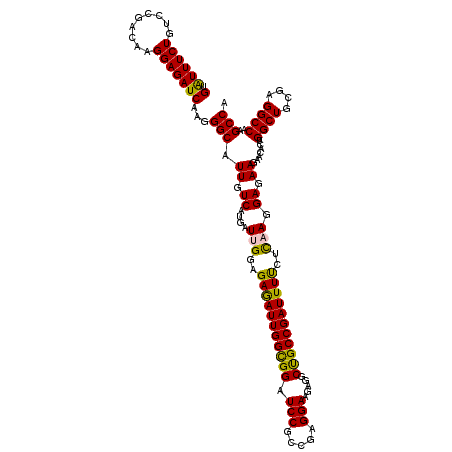
>3R_DroMel_CAF1 15287894 120 - 27905053 GUGUUUCUGUCCGACAAGGAGAUCAAGGGCAUUGUCAUGAUUGGAGAGAUUGGCGGAUCCGCCGAGGAAGAGGCUGCCGAUUUCCUUAAGGAGAAGAACACUGGCUGCGAGGCCAAGCCA (((((..(.(((...(((((((((...((((.(.((..((((.....))))((((....))))......)).).)))))))))))))..))).)..)))))(((((....)))))..... ( -45.00) >DroSec_CAF1 40437 120 - 1 GUAUUUCUGUCCGACAAGGAGAUCAAGGGCAUUGUCAUGAUCGGAGAGAUUGGCGGAUCCGCCGAGGAAGAUGCUGCCGAUUUUCUCAAGGAGAAGAACACUGGCUGCGAGGCCAAGCCA ....((((.(((....((((((((...((((((.((..((((.....))))((((....))))...)).))))))...))))))))...)))..))))...(((((....)))))..... ( -38.50) >DroSim_CAF1 41129 120 - 1 GUAUUUCUGUCGGACAAGGAGAUCAAGGGCAUUGUCAUGAUUGGAGAAAUUGGCGGAUCCGCCGAGGAAGAUGCUGCCGAUUUUCUCAAGGAGAAGAACACUGGCUGCGAGGCCAAGCCA (.((((((.........)))))))...(((.((.((....(((..((((((((((((((..(....)..))).)))))))))))..))).)).))......(((((....))))).))). ( -36.80) >DroEre_CAF1 36529 120 - 1 GUAUUUCUUUCCGACAAGGAGAUCAAGGGCAUUGUCAUGAUCGGAGAGAUUGGCGGAUCCGCCGAGGAAGAGGCCGCCGAUUUUCUCAAGGAGAAAAACACAGGCUGCGAGGCCAAGCCA ((.(((((((((.....)))(((((..(((...))).))))).(((((((((((((.(((.....))).....))))))).))))))...)))))).))...((((....))))...... ( -40.90) >DroYak_CAF1 39699 120 - 1 GUAUUUCUGUCCGACAAGGAGAUCAAGGGCAUUGUCAUGAUUGGAGAGAUUGGUGGAUCCGCCGAGGAAGAAGCCGCCGAUUUCCUCAAGGAGAAGAACACAGGCUGCGAGGCCAAGCCA ((.((..(.((((((((..............)))))....(((..(((((((((((.(((.....))).....)))))))))))..)))))).)..)).)).((((....))))...... ( -38.84) >consensus GUAUUUCUGUCCGACAAGGAGAUCAAGGGCAUUGUCAUGAUUGGAGAGAUUGGCGGAUCCGCCGAGGAAGAGGCUGCCGAUUUUCUCAAGGAGAAGAACACUGGCUGCGAGGCCAAGCCA (.((((((.........)))))))...(((.((.((....(((..(((((((((((.(((.....))).....)))))))))))..))).)).)).......((((....))))..))). (-36.00 = -35.28 + -0.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:54 2006