| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,285,433 – 15,285,574 |
| Length | 141 |
| Max. P | 0.990323 |

| Location | 15,285,433 – 15,285,543 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 85.58 |
| Mean single sequence MFE | -20.92 |
| Consensus MFE | -18.12 |
| Energy contribution | -18.32 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.69 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
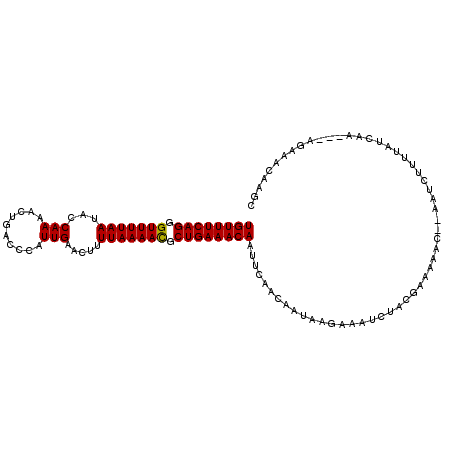
>3R_DroMel_CAF1 15285433 110 + 27905053 UUUUAUUUCCAAUUC-AUU-UAAACAAAUUGUUACUUGUUUCAGGGUUUUAAUACCAAAACUGACCCAUUGAACCUUUAAAACGCUGAAACAAUUCAACAAUAAGAAAUGUA ..............(-(((-(......((((((..(((((((((.(((((((........................))))))).)))))))))...))))))...))))).. ( -21.06) >DroSec_CAF1 37955 110 + 1 UUUUAUUUCCAAUUC-AUU-UAAACAAAUUGUUACUUGUUUCAGUGUUUUAAUGCCAAAACUGACCCAUUGAACUUUUAAAACGCUGAAACAAUUCAACAAUAAGAAAUCUA ...............-...-.......((((((..(((((((((((((((((........................)))))))))))))))))...)))))).......... ( -22.26) >DroSim_CAF1 38616 110 + 1 UUUUAGUUUCAAUUC-AUU-UAAACAAAUUGUUACUUGUUUCAGUGUUUUAAUGCCAAAACUGACCCAUUGAACUUUUAAAACGCUGAAACAAUUCAACAAUAAGAAAUCUA ...............-...-.......((((((..(((((((((((((((((........................)))))))))))))))))...)))))).......... ( -22.26) >DroEre_CAF1 34102 110 + 1 U-UUAUUUCCAAUUCCAUU-CAACAAAAUUGUUACUUGUUUCAGGGUUUUAAUACCAAAACUGACCCAUUGAACUUUUAAAACGCUGAAACAAUUUCACAAUAGCCAAUCUU .-.................-.......(((((...(((((((((.(((((((........................))))))).)))))))))....))))).......... ( -18.76) >DroYak_CAF1 37132 111 + 1 UUUUAUUUCGAAUUCCAUU-UUAAAAAAUUGUCACUUGUUUCAGGGUUUUAAUACCAAAACUGACCCAUUGAACAUUUAAAACGCUGAAACAAUUUUACAAUAGACAAUCUU ...................-.......(((((...(((((((((.(((((((........................))))))).)))))))))....))))).......... ( -18.76) >DroPer_CAF1 49042 99 + 1 UUUU-------GUUC-UUUAUAAAACAUUUGUAACUUGUUUCAGGGUUUUAA-ACCAAAACUGACCCAUUGAACUCUUAAAAUCCUGAAACAAUUCUACAA----AAAUUUA ((((-------((..-...))))))..((((((..(((((((((((((((((-.......................)))))))))))))))))...)))))----)...... ( -22.40) >consensus UUUUAUUUCCAAUUC_AUU_UAAAAAAAUUGUUACUUGUUUCAGGGUUUUAAUACCAAAACUGACCCAUUGAACUUUUAAAACGCUGAAACAAUUCAACAAUAAGAAAUCUA ...........................((((((..(((((((((.(((((((...(((..........))).....))))))).)))))))))...)))))).......... (-18.12 = -18.32 + 0.20)



| Location | 15,285,433 – 15,285,543 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 85.58 |
| Mean single sequence MFE | -23.22 |
| Consensus MFE | -19.05 |
| Energy contribution | -19.55 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15285433 110 - 27905053 UACAUUUCUUAUUGUUGAAUUGUUUCAGCGUUUUAAAGGUUCAAUGGGUCAGUUUUGGUAUUAAAACCCUGAAACAAGUAACAAUUUGUUUA-AAU-GAAUUGGAAAUAAAA ..((((((..(((((((..(((((((((.(((((((....((((..........))))..))))))).))))))))).)))))))..)...)-)))-).............. ( -23.20) >DroSec_CAF1 37955 110 - 1 UAGAUUUCUUAUUGUUGAAUUGUUUCAGCGUUUUAAAAGUUCAAUGGGUCAGUUUUGGCAUUAAAACACUGAAACAAGUAACAAUUUGUUUA-AAU-GAAUUGGAAAUAAAA ...(((((...........(((((((((.(((((((.....(....)(((......))).))))))).)))))))))....(((((..(...-.).-.)))))))))).... ( -23.50) >DroSim_CAF1 38616 110 - 1 UAGAUUUCUUAUUGUUGAAUUGUUUCAGCGUUUUAAAAGUUCAAUGGGUCAGUUUUGGCAUUAAAACACUGAAACAAGUAACAAUUUGUUUA-AAU-GAAUUGAAACUAAAA (((((.....(((((((..(((((((((.(((((((.....(....)(((......))).))))))).))))))))).)))))))..)))))-...-............... ( -22.60) >DroEre_CAF1 34102 110 - 1 AAGAUUGGCUAUUGUGAAAUUGUUUCAGCGUUUUAAAAGUUCAAUGGGUCAGUUUUGGUAUUAAAACCCUGAAACAAGUAACAAUUUUGUUG-AAUGGAAUUGGAAAUAA-A ....(..((.(((((....(((((((((.(((((((....((((..........))))..))))))).)))))))))...)))))...))..-)................-. ( -24.00) >DroYak_CAF1 37132 111 - 1 AAGAUUGUCUAUUGUAAAAUUGUUUCAGCGUUUUAAAUGUUCAAUGGGUCAGUUUUGGUAUUAAAACCCUGAAACAAGUGACAAUUUUUUAA-AAUGGAAUUCGAAAUAAAA (((((((((..........(((((((((.(((((((....((((..........))))..))))))).)))))))))..)))))))))....-................... ( -22.10) >DroPer_CAF1 49042 99 - 1 UAAAUUU----UUGUAGAAUUGUUUCAGGAUUUUAAGAGUUCAAUGGGUCAGUUUUGGU-UUAAAACCCUGAAACAAGUUACAAAUGUUUUAUAAA-GAAC-------AAAA ......(----((((((..((((((((((.(((((((...((((..........)))))-)))))).)))))))))).)))))))((((((....)-))))-------)... ( -23.90) >consensus UAGAUUUCUUAUUGUUGAAUUGUUUCAGCGUUUUAAAAGUUCAAUGGGUCAGUUUUGGUAUUAAAACCCUGAAACAAGUAACAAUUUGUUUA_AAU_GAAUUGGAAAUAAAA ..........((((((...(((((((((.(((((((....((((..........))))..))))))).)))))))))..))))))........................... (-19.05 = -19.55 + 0.50)


| Location | 15,285,467 – 15,285,574 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 84.66 |
| Mean single sequence MFE | -17.60 |
| Consensus MFE | -14.14 |
| Energy contribution | -14.00 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15285467 107 + 27905053 UGUUUCAGGGUUUUAAUACCAAAACUGACCCAUUGAACCUUUAAAACGCUGAAACAAUUCAACAAUAAGAAAUGUACUAAAAAC--AAUCUUUUAUCAA---AGAAACAAGC ((((((((.(((((((........................))))))).))))))))............................--..(((((....))---)))....... ( -16.86) >DroSec_CAF1 37989 107 + 1 UGUUUCAGUGUUUUAAUGCCAAAACUGACCCAUUGAACUUUUAAAACGCUGAAACAAUUCAACAAUAAGAAAUCUACGAAAAAC--AAUCUUUAAUCAA---AGAAACAAGC ((((((((((((((((........................))))))))))))))))............................--..(((((....))---)))....... ( -19.26) >DroSim_CAF1 38650 107 + 1 UGUUUCAGUGUUUUAAUGCCAAAACUGACCCAUUGAACUUUUAAAACGCUGAAACAAUUCAACAAUAAGAAAUCUACUAAAAAC--AAUCUCUUAUCAA---AGAAACAAGC ((((((((((((((((........................)))))))))))))))).(((....((((((..............--....))))))...---.)))...... ( -19.73) >DroEre_CAF1 34136 105 + 1 UGUUUCAGGGUUUUAAUACCAAAACUGACCCAUUGAACUUUUAAAACGCUGAAACAAUUUCACAAUAGCCAAUCUUCAAACAAC----UUUUUUAUCAA---AGAAACAAGC ((((((((.(((((((........................))))))).))))))))...........................(----(((......))---))........ ( -15.66) >DroYak_CAF1 37167 110 + 1 UGUUUCAGGGUUUUAAUACCAAAACUGACCCAUUGAACAUUUAAAACGCUGAAACAAUUUUACAAUAGACAAUCUUCGAAAAAC--AAUUCUUUAUUAAACAAGAAACAAGC ((((((((.(((((((........................))))))).))))))))............................--..(((((........)))))...... ( -16.86) >DroPer_CAF1 49070 103 + 1 UGUUUCAGGGUUUUAA-ACCAAAACUGACCCAUUGAACUCUUAAAAUCCUGAAACAAUUCUACAA----AAAUUUACCAGAAAUAUAAUUUAUAAU-AC---UAAAACAAAA ((((((((((((((((-.......................)))))))))))))))).........----...........................-..---.......... ( -17.20) >consensus UGUUUCAGGGUUUUAAUACCAAAACUGACCCAUUGAACUUUUAAAACGCUGAAACAAUUCAACAAUAAGAAAUCUACGAAAAAC__AAUCUUUUAUCAA___AGAAACAAGC ((((((((.(((((((...(((..........))).....))))))).))))))))........................................................ (-14.14 = -14.00 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:51 2006