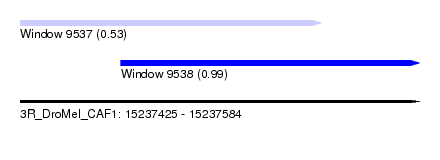
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,237,425 – 15,237,584 |
| Length | 159 |
| Max. P | 0.986867 |

| Location | 15,237,425 – 15,237,545 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.10 |
| Mean single sequence MFE | -33.60 |
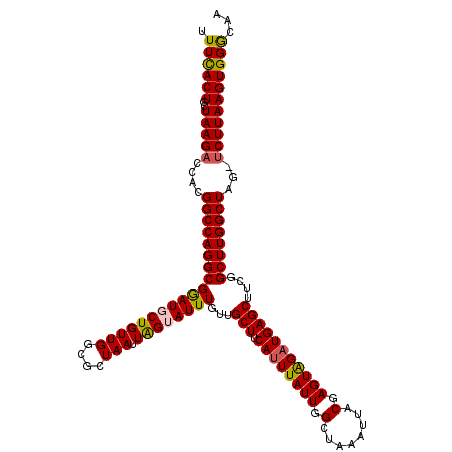
| Consensus MFE | -27.54 |
| Energy contribution | -27.46 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.82 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15237425 120 + 27905053 ACUGUUGCGCUUGGCCAUUUACACAUUGGUUUUGGCCCUGUUUCACUGCUAAGACCAUGGCCAGGCGGAUGCUGUUGGCGCUAAUUAGUAUUUGUUGCUUCAGUUAUUGGCUAAAUUACG ((((..(((((((((((.........((((((((((..((...))..)))))))))))))))))))(((((((((((....))).))))))))...))..))))................ ( -38.90) >DroSec_CAF1 4692 117 + 1 -CUGUUG--CUUGGCCAUUUACACAUUGGUUUUGGCCCUGUUUCACUGCUAAGACCACGGCCAGGCGGAUGCUGUUGGCGCUAAUUAGUAUUUGUUGCUUCAUUUAUUGGCUAAAUUACG -...(((--(((((((..........((((((((((..((...))..)))))))))).))))))))))..((.....))((((((.((((.....))))......))))))......... ( -33.50) >DroSim_CAF1 4720 118 + 1 ACUGUUG--CUUGGCCAUUUACACAUUGGUUUUGGCCCUGUUUCACUGCUAAGACCACGGCCAGGCGGAUGCUGUUGGCGCUAAUUAGUAUUUGUUGCUUCAUUUAUUGGCUAAAUUACG ....(((--(((((((..........((((((((((..((...))..)))))))))).))))))))))..((.....))((((((.((((.....))))......))))))......... ( -33.50) >DroEre_CAF1 2546 118 + 1 ACUGUUG--CUUGGCCAUUUCUACAUCGGUUUUGGCCCUGUUUCACUGCUAAGAGUACGGCCAGGCGGAUUCUGUUGCCGCUAAUUGGUAUUUGUUGCUUCAUUUAUUGGCUAAAGUACG ..((..(--(..(((((...((.....))...)))))..))..)).........((((((((((((((.........)))))....((((.....))))........)))))...)))). ( -29.70) >DroYak_CAF1 2515 118 + 1 ACUGUUG--CUUGGCCAUUUCUACAUCGGUUUUGGCCCUGUUUUACUGCUAAGACCACGGCCAGGCGAAUUCUGUUGGCGCUAAUUGGUAUUUGUUGCUUCAUUUAUUGGCCACAUUACG ....(((--(((((((((......)).(((((((((...(.....).)))))))))..))))))))))....(((.(((..(((..((((.....))))....)))...))))))..... ( -32.40) >consensus ACUGUUG__CUUGGCCAUUUACACAUUGGUUUUGGCCCUGUUUCACUGCUAAGACCACGGCCAGGCGGAUGCUGUUGGCGCUAAUUAGUAUUUGUUGCUUCAUUUAUUGGCUAAAUUACG ..........(((((((..........(((((((((..((...))..)))))))))..(((..((((((((((((((....))).))))))))))))))........)))))))...... (-27.54 = -27.46 + -0.08)

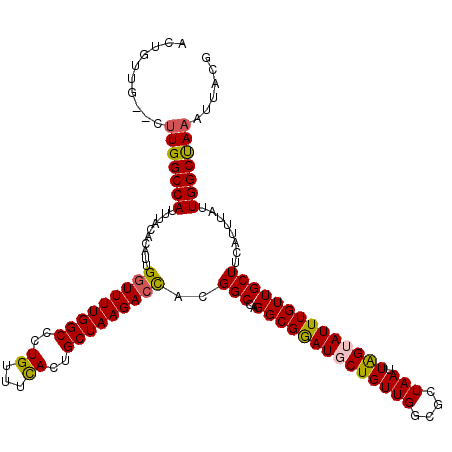
| Location | 15,237,465 – 15,237,584 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.46 |
| Mean single sequence MFE | -37.62 |
| Consensus MFE | -34.14 |
| Energy contribution | -33.98 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15237465 119 + 27905053 UUUCACUGCUAAGACCAUGGCCAGGCGGAUGCUGUUGGCGCUAAUUAGUAUUUGUUGCUUCAGUUAUUGGCUAAAUUACGAGUAGAUGAGCUUCAGCUUGGCUAG-UCUUAAGUGGGCAA .((((((..((((((..((((((((((((((((((((....))).))))))))...(((.((.(((((.(........).))))).)))))....))))))))))-)))))))))))... ( -36.80) >DroSec_CAF1 4729 119 + 1 UUUCACUGCUAAGACCACGGCCAGGCGGAUGCUGUUGGCGCUAAUUAGUAUUUGUUGCUUCAUUUAUUGGCUAAAUUACGAGUGGAUGAGCUUCGGCUUGGCUAG-UCUUAAGUGGGCAA .((((((..((((((...((((((((.((.((....((((.(((.......))).))))(((((((((.(........).))))))))))).)).)))))))).)-)))))))))))... ( -39.70) >DroSim_CAF1 4758 119 + 1 UUUCACUGCUAAGACCACGGCCAGGCGGAUGCUGUUGGCGCUAAUUAGUAUUUGUUGCUUCAUUUAUUGGCUAAAUUACGAGUGGAUGAGCUUCGGCUUGGCUAG-UCUUAAGUGGGCAA .((((((..((((((...((((((((.((.((....((((.(((.......))).))))(((((((((.(........).))))))))))).)).)))))))).)-)))))))))))... ( -39.70) >DroEre_CAF1 2584 119 + 1 UUUCACUGCUAAGAGUACGGCCAGGCGGAUUCUGUUGCCGCUAAUUGGUAUUUGUUGCUUCAUUUAUUGGCUAAAGUACGAGUAGAUGAGCUUCGGCUUGGCUUG-UCUUAAGUGGGGAA (..((((..(((((....((((((((.((......(((((.....)))))......(((.((((((((.(........).))))))))))).)).))))))))..-)))))))))..).. ( -38.50) >DroYak_CAF1 2553 120 + 1 UUUUACUGCUAAGACCACGGCCAGGCGAAUUCUGUUGGCGCUAAUUGGUAUUUGUUGCUUCAUUUAUUGGCCACAUUACGAGUAGAUGAGCUUCGGCUUGGCUUAAGCUUAAGUGGAGAA (((((((((((((.(((.((((.(((((((((.((((....)))).)).)))))))(((.((((((((.(........).)))))))))))...)))))))))).)))...))))))).. ( -33.40) >consensus UUUCACUGCUAAGACCACGGCCAGGCGGAUGCUGUUGGCGCUAAUUAGUAUUUGUUGCUUCAUUUAUUGGCUAAAUUACGAGUAGAUGAGCUUCGGCUUGGCUAG_UCUUAAGUGGGCAA .((((((..(((((....(((((((((((((((((((....))).))))))))...(((.((((((((.(........).)))))))))))....))))))))...)))))))))))... (-34.14 = -33.98 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:39 2006