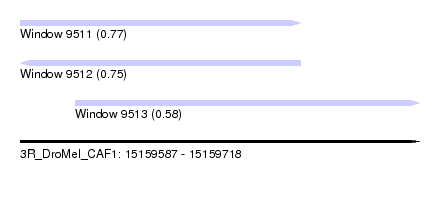
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,159,587 – 15,159,718 |
| Length | 131 |
| Max. P | 0.773595 |

| Location | 15,159,587 – 15,159,679 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 83.56 |
| Mean single sequence MFE | -26.40 |
| Consensus MFE | -17.62 |
| Energy contribution | -18.24 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
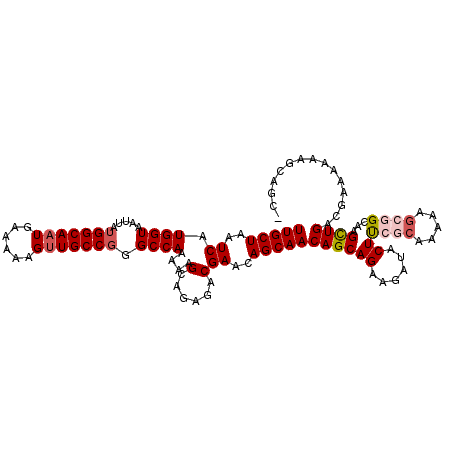
>3R_DroMel_CAF1 15159587 92 + 27905053 -----------AGC--UGGCCCCAGCAUUCAUUGCUAAUCAUGGUAAUUAUGGCAAUGAAAAAGCUGCCUGGCCAAAAAGCAGAGACGAAC--AGCAACAGCAGAAG -----------.((--(((((.((((.((((((((((.............))))))))))...))))...)))).....((..........--.))...)))..... ( -26.22) >DroSim_CAF1 82480 92 + 1 -----------AAC--UGGCCCCAGCAUUCAUUGCUAAUCAUGGUAAUUAUGGCAAUGAAAAAGUUGCCGGGCCAAAAAGCAGAGACGAAC--AGCAACAGCAGAAG -----------...--((((((((((.((((((((((.............))))))))))...))))..))))))................--.((....))..... ( -25.52) >DroEre_CAF1 81744 90 + 1 -----------AGC--UGGCCCCAGCAUUCAUUGCUAAUCAUGGUAAUUAUGGCAAUGAAAAAGUUGCCGGGCCAA-AAGC-GAGACGAGC--AGCAACAGCAGAAG -----------.((--((((((((((.((((((((((.............))))))))))...))))..)))))..-..((-.......))--)))........... ( -28.42) >DroWil_CAF1 76229 90 + 1 -----------AGUGGCAGCGACAGCAUUCAUUGCUAAUCAUGGUAAUUAUGGCAAUGAAAAAGUUGCU------AAAAGAAGAGAUACACAAAUAAACAACAGAAA -----------((..((.((....)).((((((((((.............))))))))))...))..))------................................ ( -18.02) >DroYak_CAF1 84407 91 + 1 -----------AGC--UGGCCCCAGCAUUCAUUGCUAAUCAUGGUAAUUAUGGCAAUGAAAAAGUUGCCGGGCCAAAAAGC-GAGACGAAC--AGCAACAGCAGAAG -----------.((--((((((((((.((((((((((.............))))))))))...))))..))))).....((-.........--.))...)))..... ( -27.82) >DroAna_CAF1 80172 102 + 1 GGUAGCUGGGUAGC--UGGCCCCAGCAUUCGUUGCUAAUCAUGGUAAUUAUGGCAAUGAAAAAGUUGCCAAGCCGAAAAGCA-AGACGAAC--AAAAACACCAGAAG (((.((((((....--....)))))).(((((((((..((..(((.....(((((((......))))))).)))))..))))-..))))).--......)))..... ( -32.40) >consensus ___________AGC__UGGCCCCAGCAUUCAUUGCUAAUCAUGGUAAUUAUGGCAAUGAAAAAGUUGCCGGGCCAAAAAGCAGAGACGAAC__AGCAACAGCAGAAG ................(((((.((((.((((((((((.............))))))))))...))))...)))))................................ (-17.62 = -18.24 + 0.61)


| Location | 15,159,587 – 15,159,679 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 83.56 |
| Mean single sequence MFE | -26.15 |
| Consensus MFE | -18.43 |
| Energy contribution | -19.27 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15159587 92 - 27905053 CUUCUGCUGUUGCU--GUUCGUCUCUGCUUUUUGGCCAGGCAGCUUUUUCAUUGCCAUAAUUACCAUGAUUAGCAAUGAAUGCUGGGGCCA--GCU----------- .....((.(..((.--....))..).))...((((((...((((...((((((((..(((((.....))))))))))))).)))).)))))--)..----------- ( -27.70) >DroSim_CAF1 82480 92 - 1 CUUCUGCUGUUGCU--GUUCGUCUCUGCUUUUUGGCCCGGCAACUUUUUCAUUGCCAUAAUUACCAUGAUUAGCAAUGAAUGCUGGGGCCA--GUU----------- .....((.(..((.--....))..).))...(((((((((((.....((((((((..(((((.....))))))))))))))))).))))))--)..----------- ( -27.30) >DroEre_CAF1 81744 90 - 1 CUUCUGCUGUUGCU--GCUCGUCUC-GCUU-UUGGCCCGGCAACUUUUUCAUUGCCAUAAUUACCAUGAUUAGCAAUGAAUGCUGGGGCCA--GCU----------- .....((((.....--((.......-))..-..(.(((((((.....((((((((..(((((.....)))))))))))))))))))).)))--)).----------- ( -27.80) >DroWil_CAF1 76229 90 - 1 UUUCUGUUGUUUAUUUGUGUAUCUCUUCUUUU------AGCAACUUUUUCAUUGCCAUAAUUACCAUGAUUAGCAAUGAAUGCUGUCGCUGCCACU----------- ...............................(------(((.((...((((((((..(((((.....)))))))))))))....)).)))).....----------- ( -14.90) >DroYak_CAF1 84407 91 - 1 CUUCUGCUGUUGCU--GUUCGUCUC-GCUUUUUGGCCCGGCAACUUUUUCAUUGCCAUAAUUACCAUGAUUAGCAAUGAAUGCUGGGGCCA--GCU----------- .....((((..(((--.........-(((....)))((((((.....((((((((..(((((.....))))))))))))))))))))))))--)).----------- ( -27.30) >DroAna_CAF1 80172 102 - 1 CUUCUGGUGUUUUU--GUUCGUCU-UGCUUUUCGGCUUGGCAACUUUUUCAUUGCCAUAAUUACCAUGAUUAGCAACGAAUGCUGGGGCCA--GCUACCCAGCUACC .....((((.....--((((((..-((((..((((..((((((........))))))......))..))..))))))))))((((((....--....)))))))))) ( -31.90) >consensus CUUCUGCUGUUGCU__GUUCGUCUCUGCUUUUUGGCCCGGCAACUUUUUCAUUGCCAUAAUUACCAUGAUUAGCAAUGAAUGCUGGGGCCA__GCU___________ .................................(((((((((.....((((((((..(((((.....))))))))))))))))).)))))................. (-18.43 = -19.27 + 0.83)



| Location | 15,159,605 – 15,159,718 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 91.34 |
| Mean single sequence MFE | -26.79 |
| Consensus MFE | -18.84 |
| Energy contribution | -19.84 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15159605 113 + 27905053 UUGCUAAUCAUGGUAAUUAUGGCAAUGAAAAAGCUGCCUGGCCAAAAAGCAGAGACGAACAGCAACAGCAGAAGAUACUUCGCAAAAAGCGGCAAGCUGACGAAAAAAGCAGC- (((((.....((((......((((..........))))..))))....((...........((....)).((((...)))))).......)))))((((.(.......)))))- ( -22.00) >DroSec_CAF1 83205 114 + 1 UUGCUAAUCAUGGUAAUUAUGGCAAUGAAAAAGUUGCCGGGCCAAAAAGCAGAGACGAACAGCAACAGCAGAAGAUACUUCGCAAAAAGCGGCAAGCUGACGAAAAAAGCAGCA (((((.....((((.....(((((((......))))))).))))....((...........((....)).((((...)))))).......)))))((((.(.......))))). ( -28.80) >DroSim_CAF1 82498 114 + 1 UUGCUAAUCAUGGUAAUUAUGGCAAUGAAAAAGUUGCCGGGCCAAAAAGCAGAGACGAACAGCAACAGCAGAAGAUACUUCGCAAAAAGCGGCAAGCUGACGAAAAAAGCAGCA (((((.....((((.....(((((((......))))))).))))....((...........((....)).((((...)))))).......)))))((((.(.......))))). ( -28.80) >DroEre_CAF1 81762 107 + 1 UUGCUAAUCAUGGUAAUUAUGGCAAUGAAAAAGUUGCCGGGCCAA-AAGC-GAGACGAGCAGCAACAGCAGAAGAUACUCGCCUAAAAGCAGCAAGCUGACGAAAAA--CU--- (((((.....((((.....(((((((......))))))).)))).-..((-.......)))))))((((.((......))((......)).....))))........--..--- ( -26.10) >DroYak_CAF1 84425 107 + 1 UUGCUAAUCAUGGUAAUUAUGGCAAUGAAAAAGUUGCCGGGCCAAAAAGC-GAGACGAACAGCAACAGCAGAAGAUACUCGC-AAAGAGCAGCAAGUUGACGAAAAA--CU--- (((((..((.((((.....(((((((......))))))).))))....((-(((.......((....))........)))))-...))..)))))............--..--- ( -28.26) >consensus UUGCUAAUCAUGGUAAUUAUGGCAAUGAAAAAGUUGCCGGGCCAAAAAGCAGAGACGAACAGCAACAGCAGAAGAUACUUCGCAAAAAGCGGCAAGCUGACGAAAAAAGCAGC_ (((((..((.((((.....(((((((......))))))).))))....(......)))..)))))((((((......))((((.....))))...))))............... (-18.84 = -19.84 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:05:14 2006