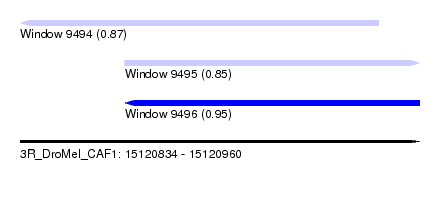
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,120,834 – 15,120,960 |
| Length | 126 |
| Max. P | 0.951295 |

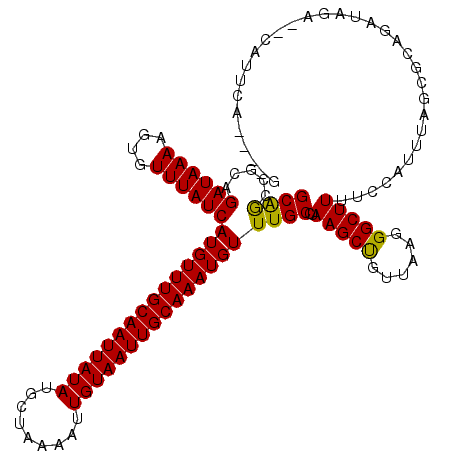
| Location | 15,120,834 – 15,120,947 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 82.36 |
| Mean single sequence MFE | -30.00 |
| Consensus MFE | -23.20 |
| Energy contribution | -22.62 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15120834 113 - 27905053 GCAGUCGCAGAUAAAAGUGUUUAUCAUGUUUGCAAUUAUAUGCUAAAAUUGUAAUUGCAAAUGUUUGCCUAAGCCGUUGAGGGCUUUUCCAUUUAGCGCAGAUAGA--CAUUCA----G ((....)).......(((((((((((((((((((((((((.........))))))))))))))).((((((((((......))))........))).)))))))))--))))..----. ( -32.50) >DroVir_CAF1 54076 111 - 1 GCGAGCGGAGAUAAAAGUGUUUAUCAUGUUUGCAAUUAUAUGCUAAAAUUGUAAUUGCAAAUGUUUGCCCAAGCUGUUAAGGGCUUUACU------CGCAGCUAAA--CCUUCAACCAG (((((..((((((((....))))))(((((((((((((((.........)))))))))))))))..((((..........))))))..))------))).......--........... ( -32.10) >DroPse_CAF1 51289 110 - 1 GCAGCCGCAGAUAAAAGUGUUUAUCAUGUUUGCAAUUAUAUGCUAAACUUGUAAUUGCAAAUGUUUGCCUAAGCUGUUGAGGGCUUUUCCAUUUAGCGUAGA-AGC--CA--CA----G (((((.(((((((((....))))))(((((((((((((((.........))))))))))))))).)))....)))))....(((((((.(.......).)))-)))--).--..----. ( -31.90) >DroGri_CAF1 65470 113 - 1 GCGAACGGCGAUAAAAUUGUUUAUCAUGUUUGCAAUUAUAUGCUAAAAUUGUAAUUGCAAAUGUUUGCCUAAGCUGUAAAGGGCUUUACU------CUCAGAUACUGGCACACACACAG ((.....))((((((....))))))..(((((((((((((.........)))))))))))))((.((((....(((...((((.....))------))))).....)))).))...... ( -28.50) >DroWil_CAF1 35768 113 - 1 GCAGUCAGCGAUAAAAGUGUUUAUCAUGUUUGCAAUUAUAUGCUAAAAUUGUAAUUGCAAAUGUUUGCCCAAGCUGUUAAGGGCUUUUCCAUUUAGAUAGAAUAGA--CAUACU----A ...(((.((.......)).(((((((((((((((((((((.........)))))))))))))))..((((..........))))...........))))))...))--).....----. ( -26.90) >DroAna_CAF1 44212 113 - 1 GCAGCAACAGAUAAAAGUGUUUAUCAUGUUUGCAAUUAUAUGCUAAAAUUGUAAUUGCAAAUGUUUGCCUAAGCUGUUGAGGGCUUUUCCAUUUGGCGCAGAUAGA--CGUUCA----G ..................((((((((((((((((((((((.........)))))))))))))))..(((.(((((......)))))........)))...))))))--).....----. ( -28.10) >consensus GCAGCCGCAGAUAAAAGUGUUUAUCAUGUUUGCAAUUAUAUGCUAAAAUUGUAAUUGCAAAUGUUUGCCUAAGCUGUUAAGGGCUUUUCCAUUUAGCGCAGAUAGA__CAUUCA____G ((((.....((((((....))))))(((((((((((((((.........)))))))))))))))))))..(((((......)))))................................. (-23.20 = -22.62 + -0.58)

| Location | 15,120,867 – 15,120,960 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 82.77 |
| Mean single sequence MFE | -20.12 |
| Consensus MFE | -13.28 |
| Energy contribution | -13.34 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.847001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15120867 93 + 27905053 UCAACGGCUUAGGCAAACAUUUGCAAUUACAAUUUUAGCAUAUAAUUGCAAACAUGAUAAACACUUUUAUCUGCGACUGCUGUUC------ACUCUCGG ..((((((....(((....((((((((((.............))))))))))...((((((....)))))))))....)))))).------........ ( -19.22) >DroVir_CAF1 54107 91 + 1 UUAACAGCUUGGGCAAACAUUUGCAAUUACAAUUUUAGCAUAUAAUUGCAAACAUGAUAAACACUUUUAUCUCCGCUCGCUGCGCUGCCAA-------- ....((((.(((((.....((((((((((.............))))))))))...((((((....))))))...)))))....))))....-------- ( -19.92) >DroPse_CAF1 51319 82 + 1 UCAACAGCUUAGGCAAACAUUUGCAAUUACAAGUUUAGCAUAUAAUUGCAAACAUGAUAAACACUUUUAUCUGCGGCUGCUC----------------- ....(((((...(((....((((((((((...(.....)...))))))))))...((((((....))))))))))))))...----------------- ( -18.40) >DroGri_CAF1 65503 88 + 1 UUUACAGCUUAGGCAAACAUUUGCAAUUACAAUUUUAGCAUAUAAUUGCAAACAUGAUAAACAAUUUUAUCGCCGUUCGCUGCUC------GCU----- ....((((...(((.....((((((((((.............))))))))))...((((((....)))))))))....))))...------...----- ( -21.12) >DroMoj_CAF1 54071 94 + 1 UUUGCAGCUUAGGCAAACAUUUGCAAUUACAAUUUUAGCAUAUAAUUGCAAACAUGAUAAACACUUUUAUCGCCGCUCGUUGCGCCGCCAAGCA----- ......((((.(((.....((((((((((.............))))))))))...((((((....))))))(.(((.....))).)))))))).----- ( -24.22) >DroPer_CAF1 47898 82 + 1 UCAACAGCUUAGGCAAACAUUUGCAAUUACAAUUUUAGCAUAUAAUUGCAAACAUGAUAAACACUUUUAUCUGCGGCUGCUC----------------- ....(((((...(((....((((((((((.............))))))))))...((((((....))))))))))))))...----------------- ( -17.82) >consensus UCAACAGCUUAGGCAAACAUUUGCAAUUACAAUUUUAGCAUAUAAUUGCAAACAUGAUAAACACUUUUAUCUCCGCCCGCUGC_C______________ ....((((...........((((((((((.............))))))))))...((((((....)))))).......))))................. (-13.28 = -13.34 + 0.06)

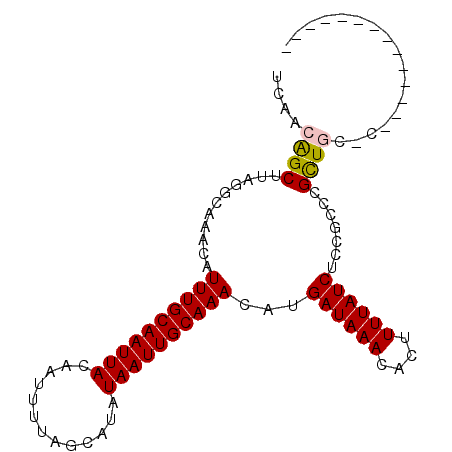
| Location | 15,120,867 – 15,120,960 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 82.77 |
| Mean single sequence MFE | -28.03 |
| Consensus MFE | -20.07 |
| Energy contribution | -20.90 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.08 |
| Mean z-score | -4.02 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15120867 93 - 27905053 CCGAGAGU------GAACAGCAGUCGCAGAUAAAAGUGUUUAUCAUGUUUGCAAUUAUAUGCUAAAAUUGUAAUUGCAAAUGUUUGCCUAAGCCGUUGA ...((.(.------.(((.((....)).((((((....))))))..(((((((((((((.........))))))))))))))))..))).......... ( -22.80) >DroVir_CAF1 54107 91 - 1 --------UUGGCAGCGCAGCGAGCGGAGAUAAAAGUGUUUAUCAUGUUUGCAAUUAUAUGCUAAAAUUGUAAUUGCAAAUGUUUGCCCAAGCUGUUAA --------.(((((((...((((((...((((((....))))))..(((((((((((((.........)))))))))))))))))))....))))))). ( -32.50) >DroPse_CAF1 51319 82 - 1 -----------------GAGCAGCCGCAGAUAAAAGUGUUUAUCAUGUUUGCAAUUAUAUGCUAAACUUGUAAUUGCAAAUGUUUGCCUAAGCUGUUGA -----------------.((((((.(((((((((....))))))(((((((((((((((.........))))))))))))))).)))....)))))).. ( -25.70) >DroGri_CAF1 65503 88 - 1 -----AGC------GAGCAGCGAACGGCGAUAAAAUUGUUUAUCAUGUUUGCAAUUAUAUGCUAAAAUUGUAAUUGCAAAUGUUUGCCUAAGCUGUAAA -----...------..(((((....(((((((((....))))))(((((((((((((((.........)))))))))))))))..)))...)))))... ( -29.20) >DroMoj_CAF1 54071 94 - 1 -----UGCUUGGCGGCGCAACGAGCGGCGAUAAAAGUGUUUAUCAUGUUUGCAAUUAUAUGCUAAAAUUGUAAUUGCAAAUGUUUGCCUAAGCUGCAAA -----......(((((((.....))(((((((((....))))))(((((((((((((((.........)))))))))))))))..)))...)))))... ( -32.30) >DroPer_CAF1 47898 82 - 1 -----------------GAGCAGCCGCAGAUAAAAGUGUUUAUCAUGUUUGCAAUUAUAUGCUAAAAUUGUAAUUGCAAAUGUUUGCCUAAGCUGUUGA -----------------.((((((.(((((((((....))))))(((((((((((((((.........))))))))))))))).)))....)))))).. ( -25.70) >consensus ______________G_GCAGCAACCGCAGAUAAAAGUGUUUAUCAUGUUUGCAAUUAUAUGCUAAAAUUGUAAUUGCAAAUGUUUGCCUAAGCUGUUAA ................(((((.......((((((....))))))(((((((((((((((.........)))))))))))))))........)))))... (-20.07 = -20.90 + 0.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:58 2006