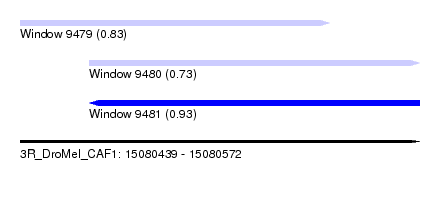
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,080,439 – 15,080,572 |
| Length | 133 |
| Max. P | 0.926962 |

| Location | 15,080,439 – 15,080,542 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 87.54 |
| Mean single sequence MFE | -23.05 |
| Consensus MFE | -16.69 |
| Energy contribution | -17.25 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15080439 103 + 27905053 CAAUGUUUUUGGAAAUGUGCAAAUGUAUUUUAUUAAUUGGCACCUGUUCUUAAACUGGUUAGCUUCCAGAGGCUAAGCCAUAAAUAUUUACUUCAUUGACCUC (((((.(((.((((..((((.(((...........))).))))...)))).))).((((((((((....))))).))))).............)))))..... ( -20.60) >DroSec_CAF1 5370 103 + 1 CAAUGUUUCUGGAAAUGCAAAAAUGUAUUUUAUUAAUUGGCACCUGUUCCUAAACGGGUUAGCUUCCAUAGGCUUAGCCAUAAAUAUUUACUUUAUUGACCUC (((((......((((((((....))))))))......(((((((((((....))))))).((((......))))..)))).............)))))..... ( -22.60) >DroSim_CAF1 3280 103 + 1 CAAUGUUUCUGUAAAUGCAGAAAUGUAUUUUAUUAAUUGGCACCUGUUCCCAAACUGGUUAGCUCCCAUGGGCUAAGCCAUAAAUAUUUACUUUAUUGACCUC ((((((((((((....))))))))(((...((((...(((((((.(((....))).)))((((((....)))))).))))..))))..)))...))))..... ( -26.20) >DroEre_CAF1 5162 102 + 1 CACUGUUUCUAGAGGCGUAGAAAUUCAAUUUAUUAAUUGGCACCUGUUCUGAAACUGGUUAGCUUUCAUAGGCU-AGCCAUAAAUAUUUAUUUUAUUGACCUC ....(((((.(.(((.((.......(((((....))))))).))).)...)))))((((((((((....)))))-)))))....................... ( -22.81) >consensus CAAUGUUUCUGGAAAUGCAGAAAUGUAUUUUAUUAAUUGGCACCUGUUCCUAAACUGGUUAGCUUCCAUAGGCUAAGCCAUAAAUAUUUACUUUAUUGACCUC (((((((((((......)))))).(((...((((...(((((((.(((....))).)))((((((....)))))).))))..))))..)))..)))))..... (-16.69 = -17.25 + 0.56)

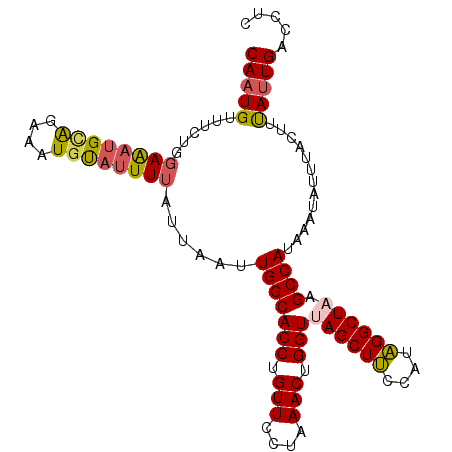
| Location | 15,080,462 – 15,080,572 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 91.97 |
| Mean single sequence MFE | -25.76 |
| Consensus MFE | -20.30 |
| Energy contribution | -20.93 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733039 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15080462 110 + 27905053 UGUAUUUUAUUAAUUGGCACCUGUUCUUAAACUGGUUAGCUUCCAGAGGCUAAGCCAUAAAUAUUUACUUCAUUGACCUCCCGUUGGCCAACAUCCCAUGGAGCUUCCAG .(((...((((...(((((((.(((....))).)))((((((....)))))).))))..))))..))).........((((...(((........))).))))....... ( -23.40) >DroSec_CAF1 5393 110 + 1 UGUAUUUUAUUAAUUGGCACCUGUUCCUAAACGGGUUAGCUUCCAUAGGCUUAGCCAUAAAUAUUUACUUUAUUGACCUCCCGUUGGCCAACAUCCCAUGGAGCUUCCAG .(((...((((...(((((((((((....))))))).((((......))))..))))..))))..))).........((((...(((........))).))))....... ( -24.30) >DroSim_CAF1 3303 110 + 1 UGUAUUUUAUUAAUUGGCACCUGUUCCCAAACUGGUUAGCUCCCAUGGGCUAAGCCAUAAAUAUUUACUUUAUUGACCUCCCGUUGGCCAACAUCCCAUGGAGCUUCCAG .............((((.........)))).((((..((((((.(((((....((((...........................))))......))))))))))).)))) ( -27.43) >DroEre_CAF1 5185 109 + 1 UUCAAUUUAUUAAUUGGCACCUGUUCUGAAACUGGUUAGCUUUCAUAGGCU-AGCCAUAAAUAUUUAUUUUAUUGACCUCCCGUUGGCCAACGUGCCAUGGAGCUUCCAU ..............((((((............((((((((((....)))))-)))))...............(((.((.......)).))).))))))(((.....))). ( -27.90) >consensus UGUAUUUUAUUAAUUGGCACCUGUUCCUAAACUGGUUAGCUUCCAUAGGCUAAGCCAUAAAUAUUUACUUUAUUGACCUCCCGUUGGCCAACAUCCCAUGGAGCUUCCAG .(((...((((...(((((((.(((....))).)))((((((....)))))).))))..))))..))).........((((...(((........))).))))....... (-20.30 = -20.93 + 0.63)

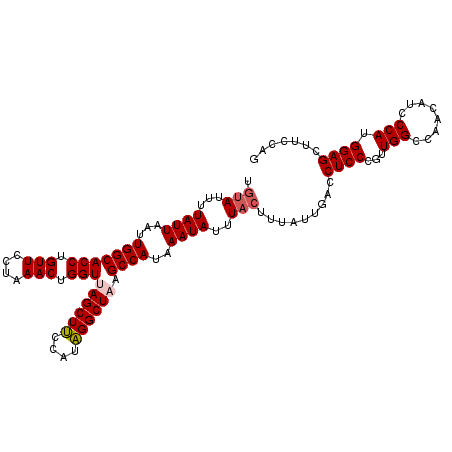
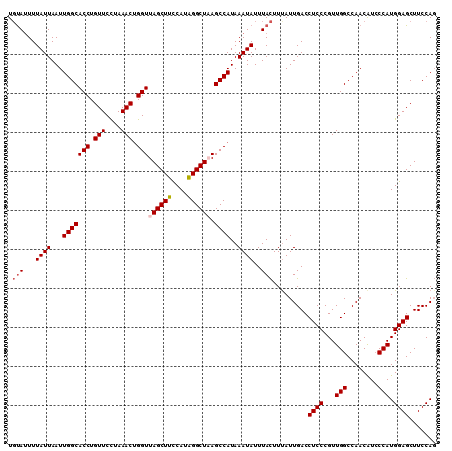
| Location | 15,080,462 – 15,080,572 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 91.97 |
| Mean single sequence MFE | -29.73 |
| Consensus MFE | -25.55 |
| Energy contribution | -26.05 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15080462 110 - 27905053 CUGGAAGCUCCAUGGGAUGUUGGCCAACGGGAGGUCAAUGAAGUAAAUAUUUAUGGCUUAGCCUCUGGAAGCUAACCAGUUUAAGAACAGGUGCCAAUUAAUAAAAUACA .(((.(((((((.(((..(((((((.......))))))).((((...........))))..))).))).)))).(((.(((....))).))).))).............. ( -25.70) >DroSec_CAF1 5393 110 - 1 CUGGAAGCUCCAUGGGAUGUUGGCCAACGGGAGGUCAAUAAAGUAAAUAUUUAUGGCUAAGCCUAUGGAAGCUAACCCGUUUAGGAACAGGUGCCAAUUAAUAAAAUACA .(((.(((((((((((...((((((..(....)....(((((.......))))))))))).))))))).)))).(((.(((....))).))).))).............. ( -31.00) >DroSim_CAF1 3303 110 - 1 CUGGAAGCUCCAUGGGAUGUUGGCCAACGGGAGGUCAAUAAAGUAAAUAUUUAUGGCUUAGCCCAUGGGAGCUAACCAGUUUGGGAACAGGUGCCAAUUAAUAAAAUACA ((((.(((((((((((.((((((((.......))))))))((((...........))))..))))).))))))..)))).((((.(.....).))))............. ( -34.20) >DroEre_CAF1 5185 109 - 1 AUGGAAGCUCCAUGGCACGUUGGCCAACGGGAGGUCAAUAAAAUAAAUAUUUAUGGCU-AGCCUAUGAAAGCUAACCAGUUUCAGAACAGGUGCCAAUUAAUAAAUUGAA .(((.....)))(((((((((((((.......)))))))..............(((.(-(((........)))).)))............)))))).............. ( -28.00) >consensus CUGGAAGCUCCAUGGGAUGUUGGCCAACGGGAGGUCAAUAAAGUAAAUAUUUAUGGCUUAGCCUAUGGAAGCUAACCAGUUUAAGAACAGGUGCCAAUUAAUAAAAUACA .(((.(((((((((((.((((((((.......))))))))..((((....)))).......))))))).)))).(((.(((....))).))).))).............. (-25.55 = -26.05 + 0.50)

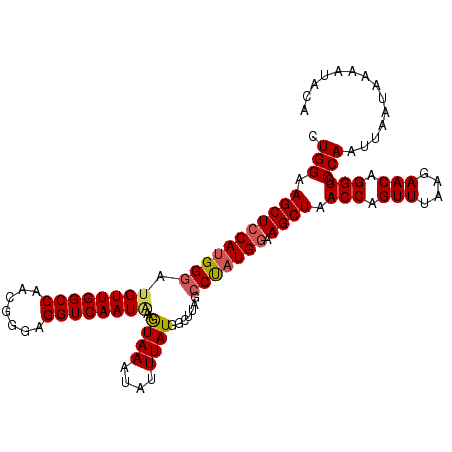
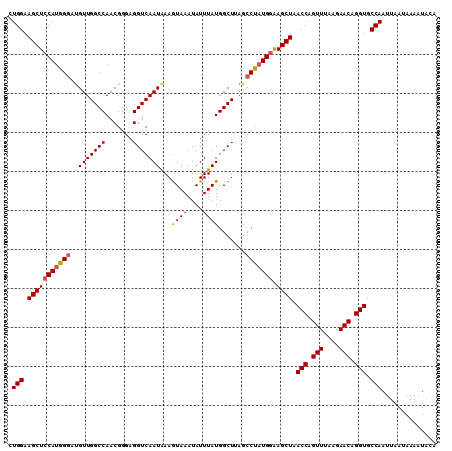
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:44 2006