
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,078,864 – 15,078,955 |
| Length | 91 |
| Max. P | 0.882046 |

| Location | 15,078,864 – 15,078,955 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 82.97 |
| Mean single sequence MFE | -22.45 |
| Consensus MFE | -13.62 |
| Energy contribution | -14.08 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.33 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
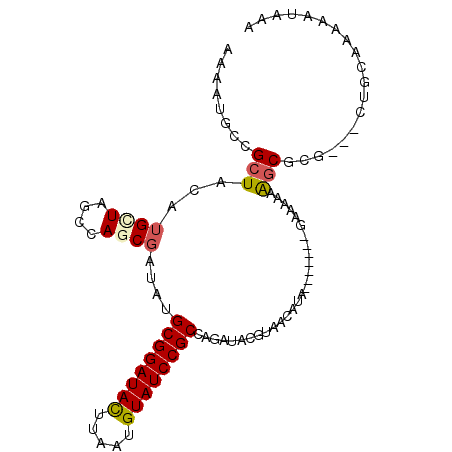
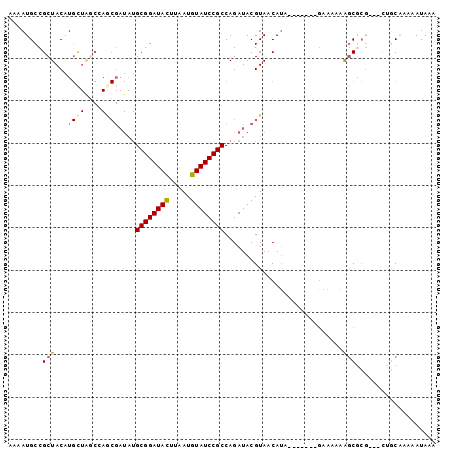
>3R_DroMel_CAF1 15078864 91 + 27905053 AAAAUGCCGCUACAUGCUAGCCAGCGAUAUGCGGAUACUUAAUGUAUCCGCCAGAUACGUAACAUA-------GAAAAAAGCGCG---CUGCAAAAAUAAA .....((((((.....(((....(((((..((((((((.....))))))))...)).)))....))-------).....)))).)---)............ ( -25.90) >DroPse_CAF1 1906 79 + 1 AAAAUGCCGCUACAU---------------GCGGAUACUUAAUGUAUCCGCCAGAUACGUAACAGA-------GACAACAACAAAAAAAAACCUAUAUAAA ...............---------------((((((((.....))))))))...............-------............................ ( -13.30) >DroSec_CAF1 3833 91 + 1 AAAAUGCCGCUACAUGUUAGCCAGCGAUAUGCGGAUACUUAAUGUAUCCGCCAGAUACGUAACAUA-------GAAAAAAGCGCG---CUGCAAAAAUAAA .....((((((.....(((....(((((..((((((((.....))))))))...)).)))....))-------).....)))).)---)............ ( -23.70) >DroSim_CAF1 1745 91 + 1 AAAAUGCCGCUACAUGCUAGCCAGCGAUAUGCGGAUACUUAAUGUAUCCGCCAGAUACGUAACAUA-------GAAAAAAGCGCG---CUGCAAAAAUAAA .....((((((.....(((....(((((..((((((((.....))))))))...)).)))....))-------).....)))).)---)............ ( -25.90) >DroEre_CAF1 3602 91 + 1 AAAAUGCCGCUACAUGCUGGCCAACGAUAUGCGGAUAUUUAAUGUAUCCGCCAGAUACGUAACAUA-------GAAAAAAGCGCG---CUGCAAAAAUAAA .....((((((.....(((....(((((..((((((((.....))))))))...)).)))....))-------).....)))).)---)............ ( -23.30) >DroAna_CAF1 2000 98 + 1 AAAAUGCCGCUACAUGCUAUCGAACGAUAUGCGGAUACUUAAUGUAUCCGCCAGAUACGUAACAUAUUGUAGAAAAAAAGGCCCG---CAACGAAAAUAAA .....(((.(((((...(((...(((((..((((((((.....))))))))...)).)))...))).))))).......)))...---............. ( -22.60) >consensus AAAAUGCCGCUACAUGCUAGCCAGCGAUAUGCGGAUACUUAAUGUAUCCGCCAGAUACGUAACAUA_______GAAAAAAGCGCG___CUGCAAAAAUAAA ........(((...((((....))))....((((((((.....))))))))............................)))................... (-13.62 = -14.08 + 0.46)

| Location | 15,078,864 – 15,078,955 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 82.97 |
| Mean single sequence MFE | -24.83 |
| Consensus MFE | -18.82 |
| Energy contribution | -19.17 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
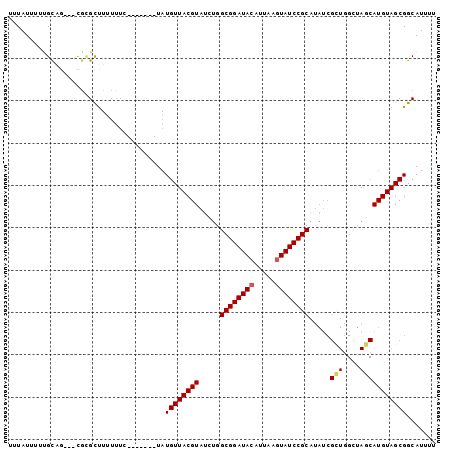
>3R_DroMel_CAF1 15078864 91 - 27905053 UUUAUUUUUGCAG---CGCGCUUUUUUC-------UAUGUUACGUAUCUGGCGGAUACAUUAAGUAUCCGCAUAUCGCUGGCUAGCAUGUAGCGGCAUUUU ............(---(.((((.....(-------((.((((((......((((((((.....))))))))....)).))))))).....))))))..... ( -27.00) >DroPse_CAF1 1906 79 - 1 UUUAUAUAGGUUUUUUUUUGUUGUUGUC-------UCUGUUACGUAUCUGGCGGAUACAUUAAGUAUCCGC---------------AUGUAGCGGCAUUUU ............................-------.(((((((((.....((((((((.....))))))))---------------)))))))))...... ( -20.90) >DroSec_CAF1 3833 91 - 1 UUUAUUUUUGCAG---CGCGCUUUUUUC-------UAUGUUACGUAUCUGGCGGAUACAUUAAGUAUCCGCAUAUCGCUGGCUAACAUGUAGCGGCAUUUU ............(---(.((((......-------(((((((.((.....((((((((.....)))))))).........))))))))).))))))..... ( -25.34) >DroSim_CAF1 1745 91 - 1 UUUAUUUUUGCAG---CGCGCUUUUUUC-------UAUGUUACGUAUCUGGCGGAUACAUUAAGUAUCCGCAUAUCGCUGGCUAGCAUGUAGCGGCAUUUU ............(---(.((((.....(-------((.((((((......((((((((.....))))))))....)).))))))).....))))))..... ( -27.00) >DroEre_CAF1 3602 91 - 1 UUUAUUUUUGCAG---CGCGCUUUUUUC-------UAUGUUACGUAUCUGGCGGAUACAUUAAAUAUCCGCAUAUCGUUGGCCAGCAUGUAGCGGCAUUUU ........(((((---....))......-------..((((((((..((((((((((.......))))).((......))))))).))))))))))).... ( -21.70) >DroAna_CAF1 2000 98 - 1 UUUAUUUUCGUUG---CGGGCCUUUUUUUCUACAAUAUGUUACGUAUCUGGCGGAUACAUUAAGUAUCCGCAUAUCGUUCGAUAGCAUGUAGCGGCAUUUU .........((((---((((........)))...((((((((((......((((((((.....))))))))........)).)))))))).)))))..... ( -27.04) >consensus UUUAUUUUUGCAG___CGCGCUUUUUUC_______UAUGUUACGUAUCUGGCGGAUACAUUAAGUAUCCGCAUAUCGCUGGCUAGCAUGUAGCGGCAUUUU .....................................((((((((.....((((((((.....)))))))).....(((....)))))))))))....... (-18.82 = -19.17 + 0.35)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:41 2006