
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,076,832 – 15,077,027 |
| Length | 195 |
| Max. P | 0.920237 |

| Location | 15,076,832 – 15,076,930 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.29 |
| Mean single sequence MFE | -29.85 |
| Consensus MFE | -17.44 |
| Energy contribution | -17.42 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.58 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15076832 98 + 27905053 CACAGAA----UAUGCGCAAGACAUUGAUUUUCGCCAGGUAAUGAGGCAACUCAUUAAUCAUUU-UUUCGUUGCG----------------CACUGGCUGGGGGAUUAUGUUGCU-GUUG .((((((----(((((....).)).(((((..((((((.(((((((....))))))).......-....(.....----------------).)))))..)..))))))))).))-)).. ( -27.80) >DroVir_CAF1 4040 97 + 1 CAGAGCA----UAUGCGC--GACAUUGAUUUUCGCCAAGUAAUGAGGCAACUCAUUAAUCAUUUUUUUAGUUGUG----------------CAUCGGCUGGGGGAUUAUGUUGCC-GGCG .......----.((((((--(((.(((........))).(((((((....)))))))............))))))----------------)))..(((((.((......)).))-))). ( -30.00) >DroGri_CAF1 4335 112 + 1 CACAGAA----UAUGUGC--GACAUUGAUUUUCGUAAAGUAAUGAGGCAACUCAUUAAUCAUUU-UUUAGUUGUGUAUGUGUGUAAAAAUUCAUCGUCAGGGGGAUUAUGUUGCU-AGCA .(((...----..)))((--(((((.((((..(......(((((((....))))))).......-.....(((((.(((.((......)).))))).))))..))))))))))).-.... ( -25.60) >DroEre_CAF1 1581 98 + 1 CACGGAA----UAUGCGCAAGACAUUGAUUUUCGCCAGGUAAUGAGGCAACUCAUUAAUCAUUU-UUUCGUUGCG----------------CACUGGCUGGGGGAUUAUGUUGCU-UUUG .......----.....(((..((((.((((..((((((.(((((((....))))))).......-....(.....----------------).)))))..)..))))))))))).-.... ( -26.60) >DroMoj_CAF1 3977 102 + 1 GGGCGCAGUGCAAUGCGC--GACAUUGAUUUUCGCGAAGUAAUGAGGCAACUCAUUAAUCAUUUUUUUAGCUGUG----------------CAUCGGCUGGGGGAUUAUGCUGCCCGGCA ..(((((((......(((--((.........)))))...(((((((....)))))))............))))))----------------)....((((((((......)).)))))). ( -37.90) >DroAna_CAF1 51 98 + 1 CGCAGAA----UAUGCGCAAGACAUUGAUUUUCGCCAGGUAAUGAGGCAACUCAUUAAUCAUUU-UUUCGUUGAG----------------CUCUGGCUGGGGGAUUAUGUUGCA-GUUG .(((...----..)))(((..((((.((((..((((((((((((((....))))))).(((...-......))).----------------.))))))..)..))))))))))).-.... ( -31.20) >consensus CACAGAA____UAUGCGC__GACAUUGAUUUUCGCCAAGUAAUGAGGCAACUCAUUAAUCAUUU_UUUAGUUGUG________________CACCGGCUGGGGGAUUAUGUUGCU_GGCG ................((..(((((.((((..(......(((((((....)))))))............((((.....................))))..)..)))))))))))...... (-17.44 = -17.42 + -0.03)



| Location | 15,076,868 – 15,076,959 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 82.73 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -21.15 |
| Energy contribution | -21.39 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.920237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15076868 91 + 27905053 AAUGAGGCAACUCAUUAAUCAUUUUUUCGUUGCGCACUGGCUGGGGGAUUAUGUUGCUGUUGCAACAAGCACACCGCCAACUGGCAA----CCG-------G ((((((....))))))............(((((.((.((((.((.(..((.((((((....))))))))..).))))))..))))))----)..-------. ( -30.90) >DroSec_CAF1 2035 91 + 1 AAUGAGGCAACUCAUUAAUCAUUUUUUCGUUGCGCACUGGCUGGGGGAUUAUGUUGCUGUUGCAACAAGCACACCGCCAACUGGCAA----CCG-------G ((((((....))))))............(((((.((.((((.((.(..((.((((((....))))))))..).))))))..))))))----)..-------. ( -30.90) >DroEre_CAF1 1617 98 + 1 AAUGAGGCAACUCAUUAAUCAUUUUUUCGUUGCGCACUGGCUGGGGGAUUAUGUUGCUUUUGCAACAAGCACACCGGCAACUCGCAA----CUGGCAACUGG ((((((....))))))............((((((.....(((((.(..((.((((((....))))))))..).)))))....)))))----).(....)... ( -34.00) >DroAna_CAF1 87 86 + 1 AAUGAGGCAACUCAUUAAUCAUUUUUUCGUUGAGCUCUGGCUGGGGGAUUAUGUUGCAGUUGCAAG-------CCGGUG--UGUCGAUGUGCCU-------G ((((((....))))))...........((((((((.((((((..(.((((.......)))).).))-------)))).)--).)))))).....-------. ( -26.10) >consensus AAUGAGGCAACUCAUUAAUCAUUUUUUCGUUGCGCACUGGCUGGGGGAUUAUGUUGCUGUUGCAACAAGCACACCGCCAACUGGCAA____CCG_______G ((((((....)))))).............((((......(((((.......((((((....))))))......))))).....))))............... (-21.15 = -21.39 + 0.25)

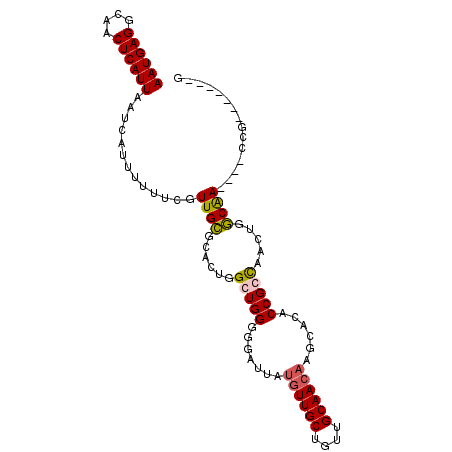

| Location | 15,076,902 – 15,076,992 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 78.52 |
| Mean single sequence MFE | -31.50 |
| Consensus MFE | -19.70 |
| Energy contribution | -20.53 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.719362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15076902 90 + 27905053 CACUGGCUGGGGGAUUAUGUUGCUGUUGCAACAAGCACACCGCCAACUGGCAACCG-------GCAACCGGCAACGAGCAACGUGCAACAGUUGCUG .....(((((((.....((((((....))))))......))(((....)))..)))-------))...((....))((((((........)))))). ( -34.40) >DroSec_CAF1 2069 72 + 1 CACUGGCUGGGGGAUUAUGUUGCUGUUGCAACAAGCACACCGCCAACUGGCAACCG-------GC--------------AACGUGCAACAGUU---- (((..(((((((.....((((((....))))))......))(((....)))..)))-------))--------------...)))........---- ( -25.20) >DroEre_CAF1 1651 97 + 1 CACUGGCUGGGGGAUUAUGUUGCUUUUGCAACAAGCACACCGGCAACUCGCAACUGGCAACUGGCGACCGGCUACGAGCAACGUGCAACAGUUGCCG .....(((((.(..((.((((((....))))))))..).)))))...........(((((((((((....((.....))....)))..)))))))). ( -34.90) >consensus CACUGGCUGGGGGAUUAUGUUGCUGUUGCAACAAGCACACCGCCAACUGGCAACCG_______GC_ACCGGC_ACGAGCAACGUGCAACAGUUGC_G ....(((((........((((((....)))))).((((...(((....))).................((....))......))))..))))).... (-19.70 = -20.53 + 0.83)

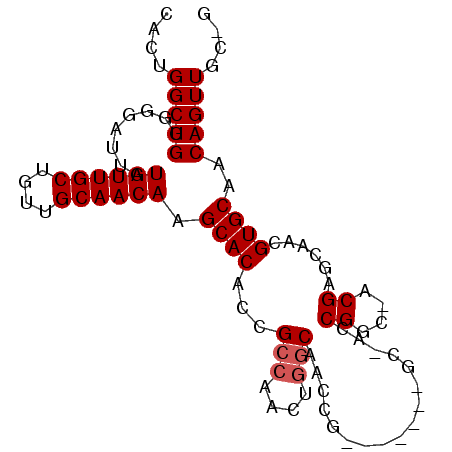

| Location | 15,076,930 – 15,077,027 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 79.34 |
| Mean single sequence MFE | -33.17 |
| Consensus MFE | -21.63 |
| Energy contribution | -22.30 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15076930 97 + 27905053 CAACAAGCACACCGCCAACUGGCAACCG-------GCAACCGGCAACGAGCAACGUGCAACAGUUGCUGCUGCCUCUGACGGAAUGCUGGCAACAUGUUGAGCG (((((((((..(((.((...((((..((-------(((((((....)).((.....))....))))))).))))..)).)))..))))(....).))))).... ( -35.60) >DroSec_CAF1 2097 77 + 1 CAACAAGCACACCGCCAACUGGCAACCG-------GC--------------AACGUGCAACAGUU------GCCUCUGACGGAAUGCUGGCAACAUGUUGAGCG (((((((((..(((((....)))....(-------((--------------(((........)))------)))......))..))))(....).))))).... ( -27.20) >DroEre_CAF1 1679 104 + 1 CAACAAGCACACCGGCAACUCGCAACUGGCAACUGGCGACCGGCUACGAGCAACGUGCAACAGUUGCCGCUGCCUCUGACGGAAUGCUGGCAACAUGUUGAGCG (((((.((((..(((....((((....(....)..)))))))((.....))...))))....((((((((..((......))...)).)))))).))))).... ( -36.70) >consensus CAACAAGCACACCGCCAACUGGCAACCG_______GC_ACCGGC_ACGAGCAACGUGCAACAGUUGC_GCUGCCUCUGACGGAAUGCUGGCAACAUGUUGAGCG (((((((((..(((.((...(((.................((....)).(((((........)))))....)))..)).)))..))))(....).))))).... (-21.63 = -22.30 + 0.67)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:39 2006