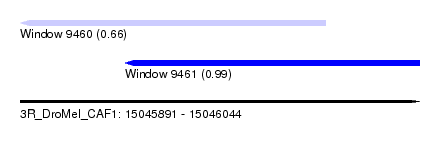
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 15,045,891 – 15,046,044 |
| Length | 153 |
| Max. P | 0.994654 |

| Location | 15,045,891 – 15,046,008 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 89.31 |
| Mean single sequence MFE | -32.34 |
| Consensus MFE | -23.66 |
| Energy contribution | -24.58 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658240 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15045891 117 - 27905053 AUUGAUUGGCUCUAGCCAAAAUUACCGAUGGUCAUUCUUGCAAGUAGUCUCAAGAUUCCUUGUUUGAAUAGGUGCUGCAUACUUUGGGGCUUGAGUUGAUUGCCUUAGAAAAUGUAA .....(((((....)))))...(((((.....)((((..(((((.((((....)))).)))))..)))).)))).(((((..((((((((...........))))))))..))))). ( -32.60) >DroSec_CAF1 48412 117 - 1 AUUGAUUGGCUCUAGCCGAAAUUUCUGAAGGUCAUUCUUGCAAGUGGUCUCAAGAUUCCUUGUUUGAAUAGAUGCUGCAUACUUUGGGGCUUGAGUUGAUUGCUUAAGACCAUGUAA .....(((((....)))))..........((((((((..(((((..(((....)))..)))))..))))....(((.((.....)).)))((((((.....))))))))))...... ( -34.50) >DroSim_CAF1 48078 117 - 1 AUUGAUUGGCUCUAGCCAAAAUUACUGAAGGUCAUUCUUGCAAGUAGUCUCAAGAUUCCUUGUUUGAAUGGGUGCUGCAUACUUUGGGGCUUGAGUUGAUUGCCUUAGACCAUGUAA .....(((((....)))))..........(.((((((..(((((.((((....)))).)))))..)))))).)..(((((..((((((((...........))))))))..))))). ( -34.20) >DroEre_CAF1 50219 116 - 1 AUUGAUUGGCCUUAGCCAAAAUUACUGAUGGUCAUUCUUGCAAUUAGUCUCAAGAUCCCUUGUUUGAAUAGGUACUGCAUACUUUGGGGCUUGAGUUCAUUGCCU-AGGCCAUGUAG .....(((((....))))).....(((((((((......(((((.((.((((((.((((..((.((...........)).))...)))))))))))).)))))..-.)))))).))) ( -31.50) >DroYak_CAF1 72020 112 - 1 GUUAAUUGGCCUUAGCCAAAA----UGAUGGUCAUUCUUGCAAUUAGUCUCAAUAUUCCUUGUUUAAAUAGGUGCUGCAUACUUUGGGGCUUGAGUUCAGUGCCU-AGACCAUGUAA .....(((((....)))))..----..((((((......((((..(((......)))..)))).....((((..(((...((((........)))).)))..)))-))))))).... ( -28.90) >consensus AUUGAUUGGCUCUAGCCAAAAUUACUGAUGGUCAUUCUUGCAAGUAGUCUCAAGAUUCCUUGUUUGAAUAGGUGCUGCAUACUUUGGGGCUUGAGUUGAUUGCCU_AGACCAUGUAA .....(((((....)))))........((((((......((((.....((((((.((((..((.((...........)).))...))))))))))....))))....)))))).... (-23.66 = -24.58 + 0.92)

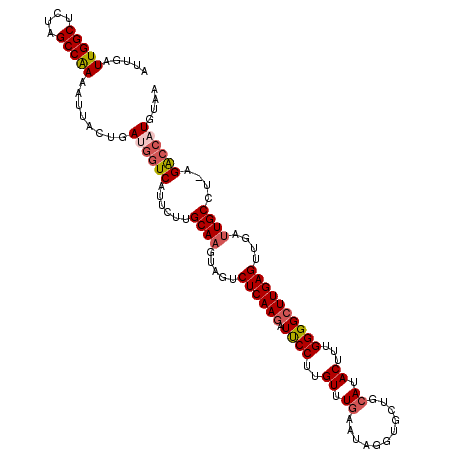
| Location | 15,045,931 – 15,046,044 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 90.09 |
| Mean single sequence MFE | -30.74 |
| Consensus MFE | -26.27 |
| Energy contribution | -27.39 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 15045931 113 - 27905053 CUGAGAUCUUAUCUCAACAAGUAUCAAUUGGCCACGAUUGAUUGGCUCUAGCCAAAAUUACCGAUGGUCAUUCUUGCAAGUAGUCUCAAGAUUCCUUGUUUGAAUAGGUGCUG .((((((...))))))...((((((....(((((((.....(((((....)))))......)).)))))((((..(((((.((((....)))).)))))..)))).)))))). ( -36.10) >DroSec_CAF1 48452 113 - 1 CUGAGAUCUUAUCUCAACAAGUAUCAAUUGGCCUGGAUUGAUUGGCUCUAGCCGAAAUUUCUGAAGGUCAUUCUUGCAAGUGGUCUCAAGAUUCCUUGUUUGAAUAGAUGCUG .((((((...))))))...((((((....((((((((....(((((....)))))....)))..)))))((((..(((((..(((....)))..)))))..)))).)))))). ( -38.50) >DroSim_CAF1 48118 113 - 1 CUGAAAUCUUAUCUCAACAAGUAUCAAUUGGCCUGGAUUGAUUGGCUCUAGCCAAAAUUACUGAAGGUCAUUCUUGCAAGUAGUCUCAAGAUUCCUUGUUUGAAUGGGUGCUG ...................((((((....(((((.((((..(((((....))))))))).....)))))((((..(((((.((((....)))).)))))..)))).)))))). ( -30.90) >DroEre_CAF1 50258 113 - 1 CUGAGAUCUUAUCUCAACAGGUAUCAAUUGGCCUCGAUUGAUUGGCCUUAGCCAAAAUUACUGAUGGUCAUUCUUGCAAUUAGUCUCAAGAUCCCUUGUUUGAAUAGGUACUG .((((((...))))))...((((((....(((((((.....(((((....)))))......))).))))((((..((((...(((....)))...))))..)))).)))))). ( -28.90) >DroYak_CAF1 72059 109 - 1 CUCAGAUCUUAUCUCAACAUGUAUCAAUUGGCCUCGGUUAAUUGGCCUUAGCCAAAA----UGAUGGUCAUUCUUGCAAUUAGUCUCAAUAUUCCUUGUUUAAAUAGGUGCUG ..(((..(((((.(.((((((((.....((((((((.....(((((....)))))..----))).)))))....))))(((......)))......)))).).)))))..))) ( -19.30) >consensus CUGAGAUCUUAUCUCAACAAGUAUCAAUUGGCCUCGAUUGAUUGGCUCUAGCCAAAAUUACUGAUGGUCAUUCUUGCAAGUAGUCUCAAGAUUCCUUGUUUGAAUAGGUGCUG .((((((...))))))...((((((....(((((.......(((((....))))).........)))))((((..(((((.((((....)))).)))))..)))).)))))). (-26.27 = -27.39 + 1.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:04:26 2006