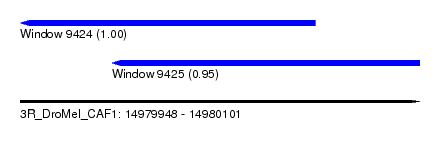
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,979,948 – 14,980,101 |
| Length | 153 |
| Max. P | 0.996060 |

| Location | 14,979,948 – 14,980,061 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 80.71 |
| Mean single sequence MFE | -28.92 |
| Consensus MFE | -21.22 |
| Energy contribution | -21.90 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.996060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14979948 113 - 27905053 GUUUAUGGUUGCCCCAUGGAUCGGGCAUCCGUAAGCGCCACUUGCUACUAAGUGGGCAAAAUAAAAAAAAUCCAAUACUUUUACAUAAUCCAUUCGAUUGGGUGUUUUUAAAA ((((((((.(((((........))))).))))))))((((((((....))))).)))......(((((.(((((((....................))))))).))))).... ( -33.35) >DroSec_CAF1 12170 99 - 1 GUUUAUGGUUGCCCAAUGGAUUGGGCAGCCGUAAGCGCUACUUGCUACUAAGUGGGCAAAAUAAA--AAAUCCAAUACUUUUACAUAAUCCAU------------UUUUAAAA ((((((((((((((((....))))))))))))))))((((((((....))))).)))........--..........................------------........ ( -31.50) >DroSim_CAF1 12263 99 - 1 GUUUAUGGUUGCCCAAUGGAUCGGGCAUCCGUAAGCGCCACUUGCUACUAAGUGGGCGAAAUAAA--AAAUCCAAUACUUUUACAUAAUCCAU------------UUUUAGAA ((((((((.(((((........))))).))))))))((((((((....))))).)))........--..........................------------........ ( -27.90) >DroEre_CAF1 12190 106 - 1 GCUUGAGGUUGCCCCUUGAAUCGGGCAGCCGUAGGCGCCA--UGCUACUAAGUGUGUAAAAUAA---AAAUCCAAUACUAUUACACAAUCCAC--AAUUGGGUGUUUUUAAAA (((((.((((((((........)))))))).)))))..((--((((....))))))(((((...---..(((((((.................--.)))))))..)))))... ( -29.97) >DroYak_CAF1 12732 108 - 1 GUUUAUGGUUGCCCGUUGAAUAGGGCAUCCUUAGGCGCCA--UGCUACUAGGUCUAUAAAAUAA---AAAUCCAAUACUAUUACACAAUCCAUUUAUUUUGGUGUUUUUAAAA (((((.((.(((((........))))).)).)))))(((.--........)))........(((---((((((((.......................)))).)))))))... ( -21.90) >consensus GUUUAUGGUUGCCCAAUGGAUCGGGCAUCCGUAAGCGCCACUUGCUACUAAGUGGGCAAAAUAAA__AAAUCCAAUACUUUUACAUAAUCCAU____UU_GGUGUUUUUAAAA ((((((((.(((((........))))).))))))))((((((((....)))))).))........................................................ (-21.22 = -21.90 + 0.68)

| Location | 14,979,983 – 14,980,101 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 86.95 |
| Mean single sequence MFE | -29.64 |
| Consensus MFE | -23.02 |
| Energy contribution | -23.70 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14979983 118 - 27905053 AUAAGUGUUAUGUUUCCUUACCCUAAUCACAGACAUCCAAGUUUAUGGUUGCCCCAUGGAUCGGGCAUCCGUAAGCGCCACUUGCUACUAAGUGGGCAAAAUAAAAAAAAUCCAAUAC .....((((.(((...........................((((((((.(((((........))))).)))))))).(((((((....)))))))))).))))............... ( -29.50) >DroSec_CAF1 12193 116 - 1 AUAAGUGUUAUCUUACCUUACCCUAAUCACAGACAUCCAAGUUUAUGGUUGCCCAAUGGAUUGGGCAGCCGUAAGCGCUACUUGCUACUAAGUGGGCAAAAUAAA--AAAUCCAAUAC .((((.((......))))))....................((((((((((((((((....))))))))))))))))((((((((....))))).)))........--........... ( -33.40) >DroSim_CAF1 12286 116 - 1 AUAAGUGUUAUCUUACCUUACCCUAAUCACAGACAUCCAAGUUUAUGGUUGCCCAAUGGAUCGGGCAUCCGUAAGCGCCACUUGCUACUAAGUGGGCGAAAUAAA--AAAUCCAAUAC .((((.((......))))))......((.(..........((((((((.(((((........))))).)))))))).(((((((....)))))))).))......--........... ( -30.20) >DroEre_CAF1 12223 113 - 1 AUAAGUGUUAUGUUACCUUACCUUAAUCGCAGACAUCCAAGCUUGAGGUUGCCCCUUGAAUCGGGCAGCCGUAGGCGCCA--UGCUACUAAGUGUGUAAAAUAA---AAAUCCAAUAC .......((((.((((...((((((...(((.........(((((.((((((((........)))))))).)))))....--)))...)))).)))))).))))---........... ( -27.62) >DroYak_CAF1 12767 113 - 1 AUAAGUGUUACGUUGCCUUAUUUUAAUCACAGGCAUCCAAGUUUAUGGUUGCCCGUUGAAUAGGGCAUCCUUAGGCGCCA--UGCUACUAGGUCUAUAAAAUAA---AAAUCCAAUAC ....(((((....((((((((((.....((.((((.(((......))).)))).)).))))))))))..(((((..((..--.))..)))))............---......))))) ( -27.50) >consensus AUAAGUGUUAUGUUACCUUACCCUAAUCACAGACAUCCAAGUUUAUGGUUGCCCAAUGGAUCGGGCAUCCGUAAGCGCCACUUGCUACUAAGUGGGCAAAAUAAA__AAAUCCAAUAC ....(((((...............................((((((((.(((((........))))).))))))))((((((((....)))))).))................))))) (-23.02 = -23.70 + 0.68)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:51 2006