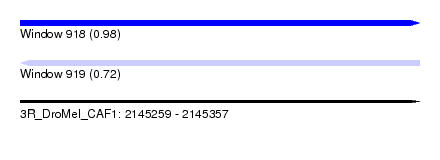
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,145,259 – 2,145,357 |
| Length | 98 |
| Max. P | 0.979216 |

| Location | 2,145,259 – 2,145,357 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 75.83 |
| Mean single sequence MFE | -25.83 |
| Consensus MFE | -14.17 |
| Energy contribution | -16.37 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2145259 98 + 27905053 CAUUGGCGGCAAUAAAAAAAAAUUUAAUAUCGAUAGGGGAGUAGCUCAUAU----GAAUUCCAAGUGUGAGUUUACACCCGGGUGGCCAAUGAAAUGCGCAU (((((((.((..((((......))))..........(((.(((((((((((----.........))))))).)))).)))..)).))))))).......... ( -28.70) >DroSec_CAF1 287952 96 + 1 CAUUGGCGGCCAUAAAA--AAAUGGAAUAUCGAUAAGGGAGUAGCUCAUAU----AAAUUCCAAGUGUGAGUUUACACCCGGGUGGCCAAUGAAAUGCGCAU (((((((.(((......--..((.(.....).))..(((.(((((((((((----.........))))))).)))).))).))).))))))).......... ( -32.40) >DroSim_CAF1 303266 95 + 1 CAUUGGCGGCCAUAAA---AAAUGGAAUAUCGAUAGGGGAGUAGCUCAUAU----GAAUUCCAAGUGUGAGUUUACACCCGGGUGGCCAAUGAAAUGCGCAU (((((((.(((.....---..((.(.....).))..(((.(((((((((((----.........))))))).)))).))).))).))))))).......... ( -32.50) >DroEre_CAF1 292079 93 + 1 CAUUGGCGGCAAUAAAA--AAAUUGAAUAUCGACAGGGGAGUAGCUUAUAC----GUAUUCCGAGUGUG---UUACACCCAGUUGGCCAAUGAAAUGCGCAU (((((((..((((....--..)))).....((((.(((..(((((.(((.(----(.....)).))).)---)))).))).))))))))))).......... ( -27.20) >DroYak_CAF1 293475 99 + 1 UAUUGGCGGCAAUAAG---AAAUGGAAUAAAAAUAAUAGAAUAGCUUAUAUAUGUAUAUUCCAAGUGUGAGUUUACACCUGGGUGGCCAAUGAAAUGCGCAU (((((((.((....((---...(((((((...(((((((......)))).)))...))))))).(((((....)))))))..)).))))))).......... ( -23.20) >DroAna_CAF1 261685 83 + 1 C--------UAAUAAACAAGGAAUGAAUGUCUAACAGGGAGUAGUCUA----------AUCC-AUUAUGAAUUCACACCCAGGUGGCCACUAUUAUGUGCAU .--------..............(((((.((......(((........----------.)))-.....)))))))(((....)))((.((......)))).. ( -11.00) >consensus CAUUGGCGGCAAUAAAA__AAAUGGAAUAUCGAUAAGGGAGUAGCUCAUAU____GAAUUCCAAGUGUGAGUUUACACCCGGGUGGCCAAUGAAAUGCGCAU (((((((..((((........))))...........(((.(((((((((((.............))))))))).)).))).....))))))).......... (-14.17 = -16.37 + 2.20)

| Location | 2,145,259 – 2,145,357 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 75.83 |
| Mean single sequence MFE | -18.99 |
| Consensus MFE | -12.88 |
| Energy contribution | -14.02 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.722624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2145259 98 - 27905053 AUGCGCAUUUCAUUGGCCACCCGGGUGUAAACUCACACUUGGAAUUC----AUAUGAGCUACUCCCCUAUCGAUAUUAAAUUUUUUUUUAUUGCCGCCAAUG ..........(((((((...((((((((......)))))))).....----...................(((((..((.....))..)))))..))))))) ( -20.00) >DroSec_CAF1 287952 96 - 1 AUGCGCAUUUCAUUGGCCACCCGGGUGUAAACUCACACUUGGAAUUU----AUAUGAGCUACUCCCUUAUCGAUAUUCCAUUU--UUUUAUGGCCGCCAAUG ..........(((((((..((.((((((......))))))(((((..----..(((((.......)))))....)))))....--......))..))))))) ( -22.40) >DroSim_CAF1 303266 95 - 1 AUGCGCAUUUCAUUGGCCACCCGGGUGUAAACUCACACUUGGAAUUC----AUAUGAGCUACUCCCCUAUCGAUAUUCCAUUU---UUUAUGGCCGCCAAUG ..........(((((((...((((((((......)))))))).....----..........................((((..---...))))..))))))) ( -21.80) >DroEre_CAF1 292079 93 - 1 AUGCGCAUUUCAUUGGCCAACUGGGUGUAA---CACACUCGGAAUAC----GUAUAAGCUACUCCCCUGUCGAUAUUCAAUUU--UUUUAUUGCCGCCAAUG ..........(((((((...(((((((...---..))))))).....----.................(.(((((........--...))))).)))))))) ( -19.70) >DroYak_CAF1 293475 99 - 1 AUGCGCAUUUCAUUGGCCACCCAGGUGUAAACUCACACUUGGAAUAUACAUAUAUAAGCUAUUCUAUUAUUUUUAUUCCAUUU---CUUAUUGCCGCCAAUA ...........((((((...((((((((......)))))))).((((....))))............................---.........)))))). ( -17.60) >DroAna_CAF1 261685 83 - 1 AUGCACAUAAUAGUGGCCACCUGGGUGUGAAUUCAUAAU-GGAU----------UAGACUACUCCCUGUUAGACAUUCAUUCCUUGUUUAUUA--------G ..........(((((..((...(((.((((((((.((((-((..----------...........)))))))).)))))).)))))..)))))--------. ( -12.42) >consensus AUGCGCAUUUCAUUGGCCACCCGGGUGUAAACUCACACUUGGAAUUC____AUAUAAGCUACUCCCCUAUCGAUAUUCAAUUU__UUUUAUUGCCGCCAAUG ..........(((((((...((((((((......))))))))...................................((((........))))..))))))) (-12.88 = -14.02 + 1.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:31 2006