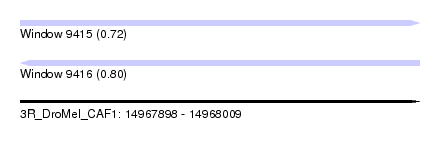
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,967,898 – 14,968,009 |
| Length | 111 |
| Max. P | 0.799476 |

| Location | 14,967,898 – 14,968,009 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.68 |
| Mean single sequence MFE | -43.45 |
| Consensus MFE | -28.87 |
| Energy contribution | -29.07 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.720525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
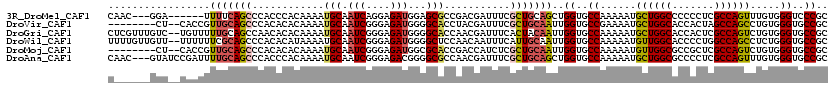
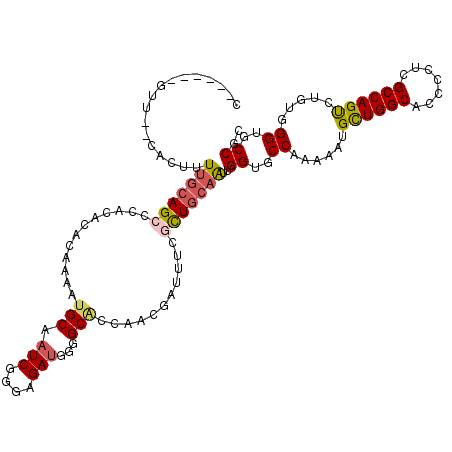
>3R_DroMel_CAF1 14967898 111 + 27905053 GCGGGACCCACAAACUGGCGAGGGGGGCCAGCAUUUUUGGCACCAGCUGCAGCGAAAUCGUCGGCGCUCCAUCUCCUGAUUGCAUUUUGUGGGUGGGCUGAAAA------UCC---GUUG ((((((((((((((...((((((((((((((.....)))))....((((..(((....))))))).......)))))..))))..)))))))))..........------)))---)).. ( -39.20) >DroVir_CAF1 311 110 + 1 GCGGCACCCACAGGCUGGCUAGUGGUGCCAGCAUUUUCGGCACCAAUUGCAGCGAAAUCGUAGGUGCCCCAUCUCCCGAUUGCAUUUUGUGUGUGGGCUGCAACGGUG--AG-------- (.((((((.((..((((.(...(((((((.........)))))))...)))))(....))).)))))).)..((((((.(((((.(..(....)..).)))))))).)--))-------- ( -43.40) >DroGri_CAF1 7729 118 + 1 GCGGCACCCACAGACUGGCGAGUGGUGCCAGCAUUUUUGGCACCAAUUGUAGUGAAAUCGUUGGUGCCCCAUCUCCCGAUUGCAUUUUGUGUGUUGGCUGCAAAAACA--GACAAACGAG ((((((((.((..((((.(((.(((((((((.....))))))))).)))))))(....))).))))))..(((....))).))..(((((.((((.........))))--.))))).... ( -39.70) >DroWil_CAF1 7802 118 + 1 GCGGCACCCAGAGGCUGGCCAGGGGUGCCAACAUUUUUGGCACCAAUUGCAAUGAAAUUGUUGGAGCCCCAUCUCCCGAUUGCAUUUUAUGUGUGGGCUGCGAAAAAA--AACAACAAAA (((((..((((...))))((.(((((.((((((.((((.(((.....)))...)))).)))))).)))))...........((((.....))))))))))).......--.......... ( -38.60) >DroMoj_CAF1 331 110 + 1 GCGGCACCCACAGACUGGCGAGCGGCGCCAACAUUUUUGGCACCAAUUGCAGCGAGAUGGUCGGUGCGCCAUCUCCCGAUUGCAUUUUGUGUGUGGGCUGCAACGGUG--AG-------- (..(((((((((.((..((((..((.(((((.....))))).))..)))).(.((((((((......)))))))).)...........)).)))))).)))..)....--..-------- ( -45.20) >DroAna_CAF1 3063 117 + 1 GCGGCACCCACAAACUGGCGAGGGGCGCCAGCAUUUUUGGCACCAGCUGCAGCGAAAUCGUUGGCGCCCCGUCUCCCGAUUGCAUUUUGUGGGUGGGCUGCAAAAUCGGAUAC---GUUG ((((((((((((((...((((((((((((((((((((..(((.....)))...))))).))))))))))).((....))))))..)))))))))..)))))............---.... ( -54.60) >consensus GCGGCACCCACAGACUGGCGAGGGGUGCCAGCAUUUUUGGCACCAAUUGCAGCGAAAUCGUUGGUGCCCCAUCUCCCGAUUGCAUUUUGUGUGUGGGCUGCAAAAACG__AAC______G ((((..((((((.((..((..(.((((((.(((((((..(((.....)))...))))).)).)))))).)(((....))).)).....)).))))))))))................... (-28.87 = -29.07 + 0.20)


| Location | 14,967,898 – 14,968,009 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.68 |
| Mean single sequence MFE | -40.38 |
| Consensus MFE | -28.04 |
| Energy contribution | -28.23 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14967898 111 - 27905053 CAAC---GGA------UUUUCAGCCCACCCACAAAAUGCAAUCAGGAGAUGGAGCGCCGACGAUUUCGCUGCAGCUGGUGCCAAAAAUGCUGGCCCCCCUCGCCAGUUUGUGGGUCCCGC ....---...------......((..((((((((...(((.((((....((.((((..........)))).)).))))))).......((((((.......))))))))))))))...)) ( -32.50) >DroVir_CAF1 311 110 - 1 --------CU--CACCGUUGCAGCCCACACACAAAAUGCAAUCGGGAGAUGGGGCACCUACGAUUUCGCUGCAAUUGGUGCCGAAAAUGCUGGCACCACUAGCCAGCCUGUGGGUGCCGC --------((--(.((((((((..............))))).))))))....((((((((((.....(((((...((((((((.......))))))))...).)))).)))))))))).. ( -46.84) >DroGri_CAF1 7729 118 - 1 CUCGUUUGUC--UGUUUUUGCAGCCAACACACAAAAUGCAAUCGGGAGAUGGGGCACCAACGAUUUCACUACAAUUGGUGCCAAAAAUGCUGGCACCACUCGCCAGUCUGUGGGUGCCGC .((((((.((--((...(((((..............))))).))))))))))((((((.................((((((((.......))))))))..(((......))))))))).. ( -36.94) >DroWil_CAF1 7802 118 - 1 UUUUGUUGUU--UUUUUUCGCAGCCCACACAUAAAAUGCAAUCGGGAGAUGGGGCUCCAACAAUUUCAUUGCAAUUGGUGCCAAAAAUGUUGGCACCCCUGGCCAGCCUCUGGGUGCCGC ..........--.......(((.((((.............(((....)))(((((((((.((((...)))).....((((((((.....))))))))..)))..)))))))))))))... ( -34.40) >DroMoj_CAF1 331 110 - 1 --------CU--CACCGUUGCAGCCCACACACAAAAUGCAAUCGGGAGAUGGCGCACCGACCAUCUCGCUGCAAUUGGUGCCAAAAAUGUUGGCGCCGCUCGCCAGUCUGUGGGUGCCGC --------..--....((.(((.((((((.((....((((...(.(((((((((...)).))))))).)))))...((((((((.....))))))))........)).))))))))).)) ( -45.60) >DroAna_CAF1 3063 117 - 1 CAAC---GUAUCCGAUUUUGCAGCCCACCCACAAAAUGCAAUCGGGAGACGGGGCGCCAACGAUUUCGCUGCAGCUGGUGCCAAAAAUGCUGGCGCCCCUCGCCAGUUUGUGGGUGCCGC ....---(((........))).((.((((((((((..((.....(....)(((((((((.((....))..(((..((....))....))))))))))))..))...))))))))))..)) ( -46.00) >consensus C______GUU__CACUUUUGCAGCCCACACACAAAAUGCAAUCGGGAGAUGGGGCACCAACGAUUUCGCUGCAAUUGGUGCCAAAAAUGCUGGCACCCCUCGCCAGUCUGUGGGUGCCGC .................(((((((............(((.(((....)))...)))...........)))))))..((..((......((((((.......)))))).....))..)).. (-28.04 = -28.23 + 0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:42 2006