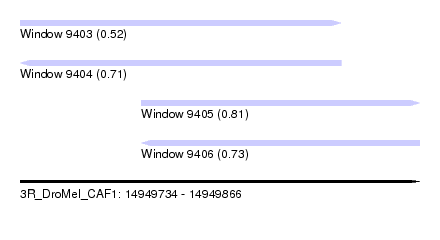
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,949,734 – 14,949,866 |
| Length | 132 |
| Max. P | 0.808410 |

| Location | 14,949,734 – 14,949,840 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 83.53 |
| Mean single sequence MFE | -34.62 |
| Consensus MFE | -23.58 |
| Energy contribution | -23.38 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14949734 106 + 27905053 UCUGUUCGCACUUCACCCAGAAUGCCUGGAAGAAGGAUUUGUGCUCGAACUGCUUCAAGUCCAAGGAUGAGCACAAGGCAGCCGCUGCA---GCAGCUGCCUCCAAAAU-- ..........((((..((((.....))))..))))...(((((((((((....)))..(((....)))))))))))((((((.((....---)).))))))........-- ( -38.60) >DroVir_CAF1 17 109 + 1 UUUGCUCGCACUUUACGCAGAAUGCGUGGAAAAAGGAUCUAUGCUCGAACUGUUUCAAGUCAAAGGAUGAGCAUAAAGCAGCCGCUGCUGCCGCAGCUGCCGCACAACA-- ..((((((..((((((((((...(((((((.......))))))).....)))).....)).))))..))))))....((.((.(((((....))))).)).))......-- ( -35.80) >DroWil_CAF1 83 106 + 1 UCUGUUCGCAUUUCACGCAAAACGCCUGGAAAAAGGAUUUGUGCUCGAAUUGCUUCAAGUCUAAGGAUGAGCACAAAGCAGCAGCUGCA---GCAGCCGCCGCCAAGAU-- .(((((.((.(((((.((.....)).)))))......((((((((((((....)))..(((....)))))))))))))))))))..((.---((....)).))......-- ( -28.80) >DroYak_CAF1 14 106 + 1 UCUGUUCGCACUUCACCCAGAAUGCCUGGAAGAAGGACUUGUGCUCGAACUGCUUCAAGUCCAAGGACGAGCACAAAGCAGCCGCUGCA---GCAGCCGCCUCCAAACU-- .......((.((((..((((.....))))..))))...(((((((((((....)))..(((....))))))))))).)).((.((((..---.)))).)).........-- ( -34.80) >DroAna_CAF1 14 106 + 1 UCUGUUCACACUUCACCCAGAAUGCCUGGAAAAAGGACCUGUGCUCCAACUGCUUCAAGUCGAAGGACGAGCAUAAGGCUGCCGCAGCA---GCAGCAGCUGCCAAGUC-- ..(((((...((((..((((.....))))......(((.((.((.......))..)).)))))))...)))))...((((((.((....---)).))))))........-- ( -32.60) >DroPer_CAF1 16 108 + 1 UUUGCUCACACUUUACGCAGAAUGCUUGGAAGAAGGAUCUGUGCUCCAACUGCUUCAAGUCCAAGGACGAGCACAAGGCAGCCGCAGCA---GCAGCCGCCUCCAAGCAGC (((((...........))))).((((((((.........((((((((....)......(((....)))))))))).(((.((.((....---)).)).))))))))))).. ( -37.10) >consensus UCUGUUCGCACUUCACCCAGAAUGCCUGGAAAAAGGAUCUGUGCUCGAACUGCUUCAAGUCCAAGGACGAGCACAAAGCAGCCGCUGCA___GCAGCCGCCGCCAAACU__ ..((((..................(((......)))...((((((((((....)))..(((....)))))))))).))))((.(((((....))))).))........... (-23.58 = -23.38 + -0.19)



| Location | 14,949,734 – 14,949,840 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.53 |
| Mean single sequence MFE | -41.82 |
| Consensus MFE | -28.17 |
| Energy contribution | -27.82 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14949734 106 - 27905053 --AUUUUGGAGGCAGCUGC---UGCAGCGGCUGCCUUGUGCUCAUCCUUGGACUUGAAGCAGUUCGAGCACAAAUCCUUCUUCCAGGCAUUCUGGGUGAAGUGCGAACAGA --..(((((((((((((((---....)))))))))))((((((......(((((......))))))))))).....((((.(((((.....))))).))))......)))) ( -42.90) >DroVir_CAF1 17 109 - 1 --UGUUGUGCGGCAGCUGCGGCAGCAGCGGCUGCUUUAUGCUCAUCCUUUGACUUGAAACAGUUCGAGCAUAGAUCCUUUUUCCACGCAUUCUGCGUAAAGUGCGAGCAAA --......(((((.(((((....))))).)))))....(((((...((.((.((((((....)))))))).))...........((((.....)))).......))))).. ( -37.90) >DroWil_CAF1 83 106 - 1 --AUCUUGGCGGCGGCUGC---UGCAGCUGCUGCUUUGUGCUCAUCCUUAGACUUGAAGCAAUUCGAGCACAAAUCCUUUUUCCAGGCGUUUUGCGUGAAAUGCGAACAGA --..((((((((((((((.---..))))))))))(((((((((.((....))...(((....)))))))))))).........)))).((((.((((...))))))))... ( -37.00) >DroYak_CAF1 14 106 - 1 --AGUUUGGAGGCGGCUGC---UGCAGCGGCUGCUUUGUGCUCGUCCUUGGACUUGAAGCAGUUCGAGCACAAGUCCUUCUUCCAGGCAUUCUGGGUGAAGUGCGAACAGA --.((((.(((((((((((---....)))))))))))(((((((((....)))..(((....)))))))))..(..((((.(((((.....))))).))))..)))))... ( -46.70) >DroAna_CAF1 14 106 - 1 --GACUUGGCAGCUGCUGC---UGCUGCGGCAGCCUUAUGCUCGUCCUUCGACUUGAAGCAGUUGGAGCACAGGUCCUUUUUCCAGGCAUUCUGGGUGAAGUGUGAACAGA --(((((((((((((((((---....))))))))....))).....((((((((......))))))))..))))))((((.(((((.....))))).))))((....)).. ( -40.10) >DroPer_CAF1 16 108 - 1 GCUGCUUGGAGGCGGCUGC---UGCUGCGGCUGCCUUGUGCUCGUCCUUGGACUUGAAGCAGUUGGAGCACAGAUCCUUCUUCCAAGCAUUCUGCGUAAAGUGUGAGCAAA (((((((((((((((((((---....)))))))))(((((((((((....)))......(....)))))))))........))))))).....((.....))...)))... ( -46.30) >consensus __AGCUUGGAGGCGGCUGC___UGCAGCGGCUGCCUUGUGCUCAUCCUUGGACUUGAAGCAGUUCGAGCACAAAUCCUUCUUCCAGGCAUUCUGCGUGAAGUGCGAACAGA .....((((.(((((((((.......)))))))))((((((((.((....))...(((....))))))))))).........))))(((((........)))))....... (-28.17 = -27.82 + -0.36)



| Location | 14,949,774 – 14,949,866 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 82.25 |
| Mean single sequence MFE | -34.33 |
| Consensus MFE | -24.16 |
| Energy contribution | -25.25 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14949774 92 + 27905053 GUGCUCGAACUGCUUCAAGUCCAAGGAUGAGCACAAGGCAGCCGCUGCAGCAGCUGCCUCCAAAAUGGGUGGCGGCCAUAAGCCGGACAAAG (((((((((....)))..(((....))))))))).(((((((.((....)).)))))))......((..((((........))))..))... ( -35.90) >DroSim_CAF1 54 92 + 1 GUGCUCGAACUGCUUCAAGUCCAAGGACGAGCACAAGGCAGCCGCUGCAGCAGCCGCCUCCAAAAUGGGUGGUGGCCAUAAGCCGGACAAGG (((((((((....)))..(((....)))))))))..(((....((....)).((((((.((......)).)))))).....)))........ ( -36.90) >DroEre_CAF1 12601 92 + 1 GUGCUCCAACUGCUUCAAGUCCAAGGACGAGCACAAGGCAGCCGCUGCAGCAGCCGCCUCCAAACUGGGUGGCGGCCAUAAGCCAGACAAAG (((((((....)......(((....)))))))))..(((....((....)).((((((.((......)).)))))).....)))........ ( -34.90) >DroWil_CAF1 123 92 + 1 GUGCUCGAAUUGCUUCAAGUCUAAGGAUGAGCACAAAGCAGCAGCUGCAGCAGCCGCCGCCAAGAUGGGCACAACACAAAAGUCAGAUGGCC (((((((...(((((...(((....))))))))....((.((.((((...)))).)).)).....))))))).........(((....))). ( -25.40) >DroYak_CAF1 54 92 + 1 GUGCUCGAACUGCUUCAAGUCCAAGGACGAGCACAAAGCAGCCGCUGCAGCAGCCGCCUCCAAACUGGGCAGCGGCCAUAAGCCAGACAAAG (((((((((....)))..(((....)))))))))......((((((((.((....))..((.....))))))))))................ ( -34.40) >DroAna_CAF1 54 89 + 1 GUGCUCCAACUGCUUCAAGUCGAAGGACGAGCAUAAGGCUGCCGCAGCAGCAGCAGCUGCCAAGUC---UGGCGGCAGCAAAGCGGACAUUC (((.(((...(((((...(((....))))))))....((((((((.(((((....)))))((....---)))))))))).....)))))).. ( -38.50) >consensus GUGCUCGAACUGCUUCAAGUCCAAGGACGAGCACAAGGCAGCCGCUGCAGCAGCCGCCUCCAAAAUGGGUGGCGGCCAUAAGCCAGACAAAG (((((((((....)))..(((....)))))))))...((((...))))....((((((.((......)).))))))................ (-24.16 = -25.25 + 1.09)



| Location | 14,949,774 – 14,949,866 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 82.25 |
| Mean single sequence MFE | -35.78 |
| Consensus MFE | -27.37 |
| Energy contribution | -28.57 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14949774 92 - 27905053 CUUUGUCCGGCUUAUGGCCGCCACCCAUUUUGGAGGCAGCUGCUGCAGCGGCUGCCUUGUGCUCAUCCUUGGACUUGAAGCAGUUCGAGCAC .......((((.....))))............(((((((((((....)))))))))))((((((......(((((......))))))))))) ( -36.90) >DroSim_CAF1 54 92 - 1 CCUUGUCCGGCUUAUGGCCACCACCCAUUUUGGAGGCGGCUGCUGCAGCGGCUGCCUUGUGCUCGUCCUUGGACUUGAAGCAGUUCGAGCAC ........(((.....))).............(((((((((((....)))))))))))(((((((((....)))..(((....))))))))) ( -38.10) >DroEre_CAF1 12601 92 - 1 CUUUGUCUGGCUUAUGGCCGCCACCCAGUUUGGAGGCGGCUGCUGCAGCGGCUGCCUUGUGCUCGUCCUUGGACUUGAAGCAGUUGGAGCAC ..(((..((((........))))..)))....(((((((((((....)))))))))))(((((((((....)))......(....))))))) ( -38.00) >DroWil_CAF1 123 92 - 1 GGCCAUCUGACUUUUGUGUUGUGCCCAUCUUGGCGGCGGCUGCUGCAGCUGCUGCUUUGUGCUCAUCCUUAGACUUGAAGCAAUUCGAGCAC ((.(((..(((......)))))).)).....(((((((((((...)))))))))))..((((((.((....))...(((....))))))))) ( -31.70) >DroYak_CAF1 54 92 - 1 CUUUGUCUGGCUUAUGGCCGCUGCCCAGUUUGGAGGCGGCUGCUGCAGCGGCUGCUUUGUGCUCGUCCUUGGACUUGAAGCAGUUCGAGCAC ........(((....(((((((((.(((((.......)))))..))))))))))))..(((((((((....)))..(((....))))))))) ( -35.20) >DroAna_CAF1 54 89 - 1 GAAUGUCCGCUUUGCUGCCGCCA---GACUUGGCAGCUGCUGCUGCUGCGGCAGCCUUAUGCUCGUCCUUCGACUUGAAGCAGUUGGAGCAC ........((...((((((((..---.....((((((....)))))))))))))).....))..((.((((((((......)))))))).)) ( -34.81) >consensus CUUUGUCCGGCUUAUGGCCGCCACCCAUUUUGGAGGCGGCUGCUGCAGCGGCUGCCUUGUGCUCGUCCUUGGACUUGAAGCAGUUCGAGCAC ........(((.....))).............(((((((((((....)))))))))))(((((((((....)))..(((....))))))))) (-27.37 = -28.57 + 1.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:32 2006