
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,949,306 – 14,949,538 |
| Length | 232 |
| Max. P | 0.964014 |

| Location | 14,949,306 – 14,949,426 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.76 |
| Mean single sequence MFE | -33.33 |
| Consensus MFE | -31.76 |
| Energy contribution | -31.45 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14949306 120 + 27905053 UCAUCAUCAGCAAAAUUUCAAUAAUUUCAUUAUCAAGAUGAAGAUUGAUUGAUAAGCUAGCUGCCUGGGCAAAUGGCAGUGUGUGGCCAUGAUUUCGGGGCUGGAUGAUGACCACAUGGC (((((((((((......(((((..(((((((.....)))))))....))))).......)))((((.((...(((((........)))))....)).))))..))))))))((....)). ( -35.52) >DroSec_CAF1 85006 116 + 1 UCAUCAUCGGCAAAAUUUCAAUAAUUCCAUUAUCAAGAUGAAGAUUGAUUGAUAAG----CUGCCUGGGCAAAUGGCAGUGUGUGGCCAUGAUUUCGGGGCUGGAUGAUGACCACAUGGC (((((((((((......(((((...((((((.....))))..))...)))))...)----))((((.((...(((((........)))))....)).))))..))))))))((....)). ( -32.30) >DroSim_CAF1 77952 116 + 1 UCAUCAUCAGCAAAAUUUCAAUAAUUCCAUUAUCAAGAUGAAGAUUGAUUGAUAAG----CUGCCUGGGCAAAUGGCAGUGUGUGGCCAUGAUUUCGGGGCUGGAUGAUGACCACAUGGC (((((((((((......(((((...((((((.....))))..))...)))))....----...((((((...(((((........)))))...))))))))).))))))))((....)). ( -32.80) >DroYak_CAF1 82043 116 + 1 UCAUCAUCAGCAAAAUUUCAAUAAUUCCAUUAUCAAGAUGAAGAUUGAUUGAUAAG----CUGGCCGGGCAAAUGGCAUUUUGUGGCCAUGAUUUCGGGGCUGGAUGAUGACCACAUGGC (((((((((((...............((((((((((............))))))).----.)))(((((...(((((........)))))...))))).))).))))))))((....)). ( -32.70) >consensus UCAUCAUCAGCAAAAUUUCAAUAAUUCCAUUAUCAAGAUGAAGAUUGAUUGAUAAG____CUGCCUGGGCAAAUGGCAGUGUGUGGCCAUGAUUUCGGGGCUGGAUGAUGACCACAUGGC (((((((((((......(((((...((((((.....))))..))...)))))...........((((((...(((((........)))))...))))))))).))))))))((....)). (-31.76 = -31.45 + -0.31)

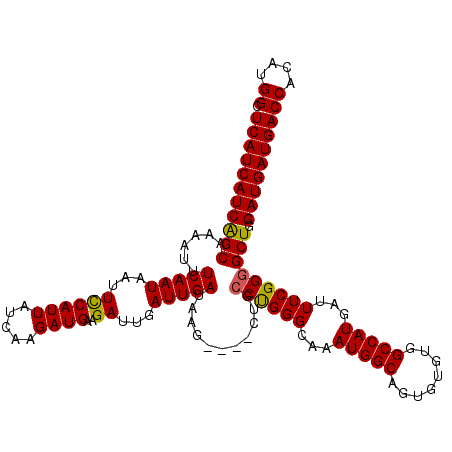
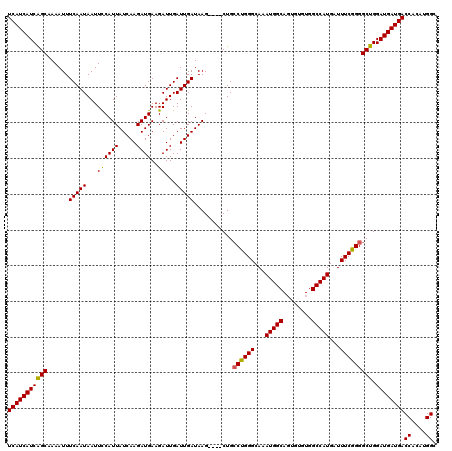
| Location | 14,949,386 – 14,949,506 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -44.70 |
| Consensus MFE | -40.22 |
| Energy contribution | -41.48 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.893057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14949386 120 + 27905053 GUGUGGCCAUGAUUUCGGGGCUGGAUGAUGACCACAUGGCCACAUGGCACAGUCGCAAUUGCUGGGCUCAACUUCUCCACUCAAUCAAUAUUGAGUCAUGCGGCAGAAGUUGAGAUUGCG (((((((((((...(((...(.....).)))...))))))))))).(((((((.......))))..(((((((((((((((((((....))))))).....)).))))))))))..))). ( -46.10) >DroSec_CAF1 85082 120 + 1 GUGUGGCCAUGAUUUCGGGGCUGGAUGAUGACCACAUGGCCACAUGGCACAGUCGCAAUUGCUGGGCUCAACUUCUCCACUCAAUCAAUAUUGAGUCAUGCGGCAGAAGUUGAGAUUGCG (((((((((((...(((...(.....).)))...))))))))))).(((((((.......))))..(((((((((((((((((((....))))))).....)).))))))))))..))). ( -46.10) >DroSim_CAF1 78028 120 + 1 GUGUGGCCAUGAUUUCGGGGCUGGAUGAUGACCACAUGGCCACAUGGCACAGUCGCAAUUGCUGGGCUCAACUUCUCCACUCAAUCAAUAUUGAGUCAUGCGGCAGAAGUUGAGAUUGCG (((((((((((...(((...(.....).)))...))))))))))).(((((((.......))))..(((((((((((((((((((....))))))).....)).))))))))))..))). ( -46.10) >DroYak_CAF1 82119 120 + 1 UUGUGGCCAUGAUUUCGGGGCUGGAUGAUGACCACAUGGCCACUUGGCAGAGUCACAAUUGCUGGGCUCAACUUCUGCACUCAAUCAAUAUUAAGUCAUGCGGCAGAAGUUGAGAUUGCG ..(((((((((...(((...(.....).)))...)))))))))(..((((........))))..).((((((((((((.(.((..(........)...)).)))))))))))))...... ( -40.50) >consensus GUGUGGCCAUGAUUUCGGGGCUGGAUGAUGACCACAUGGCCACAUGGCACAGUCGCAAUUGCUGGGCUCAACUUCUCCACUCAAUCAAUAUUGAGUCAUGCGGCAGAAGUUGAGAUUGCG (((((((((((...(((...(.....).)))...))))))))))).(((((((.......))))..(((((((((((((((((((....))))))).....)).))))))))))..))). (-40.22 = -41.48 + 1.25)

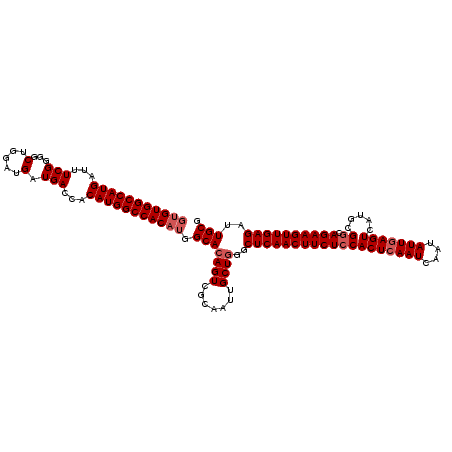
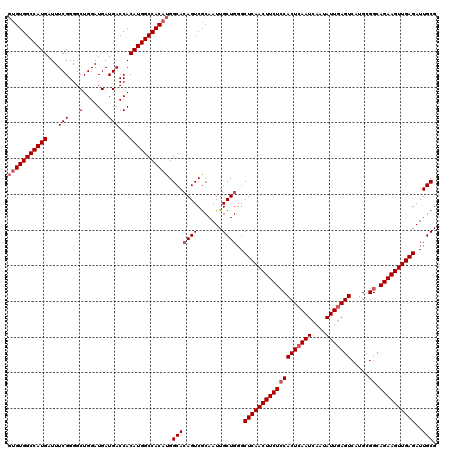
| Location | 14,949,386 – 14,949,506 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -44.18 |
| Consensus MFE | -41.56 |
| Energy contribution | -41.43 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.58 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.964014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14949386 120 - 27905053 CGCAAUCUCAACUUCUGCCGCAUGACUCAAUAUUGAUUGAGUGGAGAAGUUGAGCCCAGCAAUUGCGACUGUGCCAUGUGGCCAUGUGGUCAUCAUCCAGCCCCGAAAUCAUGGCCACAC ((((((((((((((((.((.....(((((((....))))))))))))))))))).......)))))).........((((((((((...((.............))...)))))))))). ( -44.53) >DroSec_CAF1 85082 120 - 1 CGCAAUCUCAACUUCUGCCGCAUGACUCAAUAUUGAUUGAGUGGAGAAGUUGAGCCCAGCAAUUGCGACUGUGCCAUGUGGCCAUGUGGUCAUCAUCCAGCCCCGAAAUCAUGGCCACAC ((((((((((((((((.((.....(((((((....))))))))))))))))))).......)))))).........((((((((((...((.............))...)))))))))). ( -44.53) >DroSim_CAF1 78028 120 - 1 CGCAAUCUCAACUUCUGCCGCAUGACUCAAUAUUGAUUGAGUGGAGAAGUUGAGCCCAGCAAUUGCGACUGUGCCAUGUGGCCAUGUGGUCAUCAUCCAGCCCCGAAAUCAUGGCCACAC ((((((((((((((((.((.....(((((((....))))))))))))))))))).......)))))).........((((((((((...((.............))...)))))))))). ( -44.53) >DroYak_CAF1 82119 120 - 1 CGCAAUCUCAACUUCUGCCGCAUGACUUAAUAUUGAUUGAGUGCAGAAGUUGAGCCCAGCAAUUGUGACUCUGCCAAGUGGCCAUGUGGUCAUCAUCCAGCCCCGAAAUCAUGGCCACAA ((((((((((((((((((......(((((((....))))))))))))))))))).......))))))..........(((((((((...((.............))...))))))))).. ( -43.13) >consensus CGCAAUCUCAACUUCUGCCGCAUGACUCAAUAUUGAUUGAGUGGAGAAGUUGAGCCCAGCAAUUGCGACUGUGCCAUGUGGCCAUGUGGUCAUCAUCCAGCCCCGAAAUCAUGGCCACAC ((((((((((((((((.((.....(((((((....))))))))))))))))))).......))))))..........(((((((((...((.............))...))))))))).. (-41.56 = -41.43 + -0.13)



| Location | 14,949,426 – 14,949,538 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 97.32 |
| Mean single sequence MFE | -36.83 |
| Consensus MFE | -32.33 |
| Energy contribution | -32.45 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.691928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14949426 112 + 27905053 CACAUGGCACAGUCGCAAUUGCUGGGCUCAACUUCUCCACUCAAUCAAUAUUGAGUCAUGCGGCAGAAGUUGAGAUUGCGUUGCAGGCGGCAGUGAAAGUUAAUUGUGAGCU .....(((...((((((((.((....(((((((((((((((((((....))))))).....)).))))))))))...))))))).))).(((((........)))))..))) ( -37.70) >DroSec_CAF1 85122 112 + 1 CACAUGGCACAGUCGCAAUUGCUGGGCUCAACUUCUCCACUCAAUCAAUAUUGAGUCAUGCGGCAGAAGUUGAGAUUGCGUUGCAGGCGGCAGUGAAAGUUAAUUGUGAGCU .....(((...((((((((.((....(((((((((((((((((((....))))))).....)).))))))))))...))))))).))).(((((........)))))..))) ( -37.70) >DroSim_CAF1 78068 112 + 1 CACAUGGCACAGUCGCAAUUGCUGGGCUCAACUUCUCCACUCAAUCAAUAUUGAGUCAUGCGGCAGAAGUUGAGAUUGCGUUGCAGGCGGCAGUGAAAGUUAAUUGUGAGCU .....(((...((((((((.((....(((((((((((((((((((....))))))).....)).))))))))))...))))))).))).(((((........)))))..))) ( -37.70) >DroYak_CAF1 82159 112 + 1 CACUUGGCAGAGUCACAAUUGCUGGGCUCAACUUCUGCACUCAAUCAAUAUUAAGUCAUGCGGCAGAAGUUGAGAUUGCGUUGCAGGCGGUAGUGAAAGUUAAUUGUGAGCU ((((((((....((((.((((((...((((((((((((.(.((..(........)...)).)))))))))))))..(((...))))))))).))))..)))))..))).... ( -34.20) >consensus CACAUGGCACAGUCGCAAUUGCUGGGCUCAACUUCUCCACUCAAUCAAUAUUGAGUCAUGCGGCAGAAGUUGAGAUUGCGUUGCAGGCGGCAGUGAAAGUUAAUUGUGAGCU .....(((....(((((((((((...(((((((((((((((((((....))))))).....)).))))))))))(((((((.....)).)))))...)).)))))))))))) (-32.33 = -32.45 + 0.12)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:28 2006