
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,948,266 – 14,948,654 |
| Length | 388 |
| Max. P | 0.953024 |

| Location | 14,948,266 – 14,948,383 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.12 |
| Mean single sequence MFE | -38.88 |
| Consensus MFE | -32.86 |
| Energy contribution | -32.42 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.633060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14948266 117 + 27905053 CGUGGCAGCAACAGAAGUCGAGCACUGCGGCCACAUUUAACAAUGGGUCAACUUUCCUCGGCGC---UGGCGUUGAUAAAUGUUGCCUCAAAACAAGCCAGAAACUAUAAUGUUGCUGUU ...((((((((((...((((((......((((.((((....)))))))).......)))))).(---((((.((((...........)))).....))))).........)))))))))) ( -37.32) >DroSim_CAF1 76881 117 + 1 CGUGGCAGCAACAGAAGUCGAGCACUCCGGCCACAUUUAACAAUGGGUCAACUUUCCUCGGCGC---UGGCGUUGAUAAAUGUUGCCUCAAAAUAAGCCAGAAACUGUGAUGUUGCUGUU ...((((((((((...((((((......((((.((((....)))))))).......)))))).(---((((.((((...........)))).....))))).........)))))))))) ( -37.32) >DroYak_CAF1 80950 119 + 1 CGUGGCAGCAACAGAAGUCGAGCACUUCGGCCACAUUUAACAAUGGGUCAACUUUCCUUGGCGCUGCUGGCGUUGAUAAAUGUUGCC-CAAAACAAGCCAGAAACUGUGGUGUUGCUGUU ...(((((((((.(((((.....))))).((((((........((((.((((.((..(..((((.....))))..)..)).))))))-))...(......)....))))))))))))))) ( -42.00) >consensus CGUGGCAGCAACAGAAGUCGAGCACUCCGGCCACAUUUAACAAUGGGUCAACUUUCCUCGGCGC___UGGCGUUGAUAAAUGUUGCCUCAAAACAAGCCAGAAACUGUGAUGUUGCUGUU ...((((((((((...((((((......((((.((((....)))))))).......)))))).....(((((((....)))(((.......)))..))))..........)))))))))) (-32.86 = -32.42 + -0.44)



| Location | 14,948,306 – 14,948,423 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.37 |
| Mean single sequence MFE | -38.27 |
| Consensus MFE | -34.92 |
| Energy contribution | -34.55 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915851 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14948306 117 + 27905053 CAAUGGGUCAACUUUCCUCGGCGC---UGGCGUUGAUAAAUGUUGCCUCAAAACAAGCCAGAAACUAUAAUGUUGCUGUUGCUGUCAUCUUCAUUGUUGUCGCCGCGACAGCAGCAGCAU ...((((.((((.((..((((((.---...))))))..)).)))).)))).......................(((((((((((((.....((....)).......))))))))))))). ( -36.90) >DroSec_CAF1 83999 117 + 1 CAAUGGGUCAACUUUCCUCGGCGC---UGGCGUUGAUGAAUGUUGCCUUAAAAUAAGCCAGAAACUGCGAUGUUGCUGUUGCUGUCAUCUUCAUUGUUGUCGCCGCGACAGCAGCAACAU ....(((.((((.(((.((((((.---...)))))).))).)))))))........((........))(.((((((((((((((.((.(......).)).))..)))))))))))).).. ( -38.20) >DroSim_CAF1 76921 117 + 1 CAAUGGGUCAACUUUCCUCGGCGC---UGGCGUUGAUAAAUGUUGCCUCAAAAUAAGCCAGAAACUGUGAUGUUGCUGUUGCUGUCAUCUUCAUUGUUGUCGCCGCGACAGCAGCAACAU ...((((.((((.((..((((((.---...))))))..)).)))).))))................(((.((((((((((((((.((.(......).)).))..)))))))))))).))) ( -36.70) >DroYak_CAF1 80990 119 + 1 CAAUGGGUCAACUUUCCUUGGCGCUGCUGGCGUUGAUAAAUGUUGCC-CAAAACAAGCCAGAAACUGUGGUGUUGCUGUUGCUGUCAUCUUCAUUGUUGUCGCCGCGACAGCAGCAACAU ...((((.((((.((..(..((((.....))))..)..)).))))))-))......((((.......))))(((((((((((((.((.(......).)).))..)))))))))))..... ( -41.30) >consensus CAAUGGGUCAACUUUCCUCGGCGC___UGGCGUUGAUAAAUGUUGCCUCAAAACAAGCCAGAAACUGUGAUGUUGCUGUUGCUGUCAUCUUCAUUGUUGUCGCCGCGACAGCAGCAACAU ...((((.((((.((..(((((((.....)))))))..)).)))))).))....................((((((((((((((.((.(......).)).))..)))))))))))).... (-34.92 = -34.55 + -0.37)

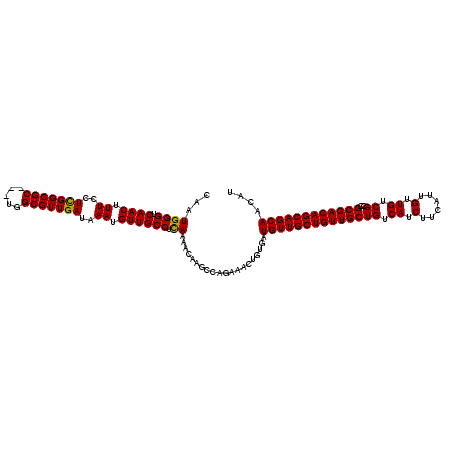

| Location | 14,948,343 – 14,948,463 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.64 |
| Mean single sequence MFE | -32.62 |
| Consensus MFE | -22.79 |
| Energy contribution | -23.10 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14948343 120 + 27905053 UGUUGCCUCAAAACAAGCCAGAAACUAUAAUGUUGCUGUUGCUGUCAUCUUCAUUGUUGUCGCCGCGACAGCAGCAGCAUGGCCAAAAACACCAGCAACAUCAGCAACACCUUCAAAUUU ((((((..........((((..(((......)))((((((((((((.....((....)).......)))))))))))).))))...........)))))).................... ( -33.30) >DroSec_CAF1 84036 111 + 1 UGUUGCCUUAAAAUAAGCCAGAAACUGCGAUGUUGCUGUUGCUGUCAUCUUCAUUGUUGUCGCCGCGACAGCAGCAACAUCGCCAAAAACACCAGC---------AACACCUUCAAAUUU ((((((((((...)))).........(((((((((((((((.(((......((....)).....))).)))))))))))))))...........))---------))))........... ( -33.40) >DroSim_CAF1 76958 111 + 1 UGUUGCCUCAAAAUAAGCCAGAAACUGUGAUGUUGCUGUUGCUGUCAUCUUCAUUGUUGUCGCCGCGACAGCAGCAACAUGGCCAAAAACCCCAGC---------AACACCUUCAAAUUU ((((((..........((((.....(((..((((((((((((((.((.(......).)).))..)))))))))))))))))))...........))---------))))........... ( -30.90) >DroYak_CAF1 81030 119 + 1 UGUUGCC-CAAAACAAGCCAGAAACUGUGGUGUUGCUGUUGCUGUCAUCUUCAUUGUUGUCGCCGCGACAGCAGCAACAUGGCCACCAACAUCAUAAACACCAGCAACACUCCCAGAUUU ((((((.-..........(((...)))(((((((..(((((..(((((.....(((((((((...)))))))))....)))))...))))).....)))))))))))))........... ( -32.90) >consensus UGUUGCCUCAAAACAAGCCAGAAACUGUGAUGUUGCUGUUGCUGUCAUCUUCAUUGUUGUCGCCGCGACAGCAGCAACAUGGCCAAAAACACCAGC_________AACACCUUCAAAUUU ((((.......)))).........(((((.((((((((((((((.((.(......).)).))..)))))))))))).)))))...................................... (-22.79 = -23.10 + 0.31)

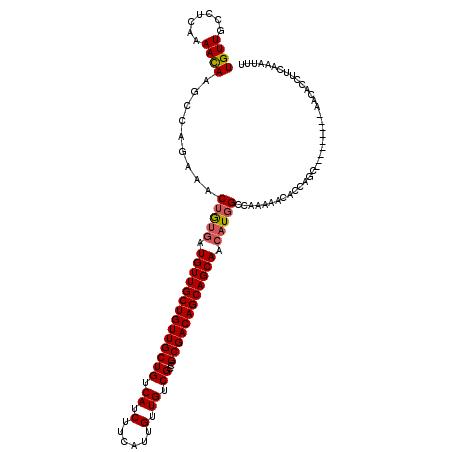

| Location | 14,948,343 – 14,948,463 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.64 |
| Mean single sequence MFE | -37.52 |
| Consensus MFE | -28.79 |
| Energy contribution | -32.23 |
| Covariance contribution | 3.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862279 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14948343 120 - 27905053 AAAUUUGAAGGUGUUGCUGAUGUUGCUGGUGUUUUUGGCCAUGCUGCUGCUGUCGCGGCGACAACAAUGAAGAUGACAGCAACAGCAACAUUAUAGUUUCUGGCUUGUUUUGAGGCAACA .........(((((((((...(((((((..(((((((....((.((((((....)))))).))....)))))))..))))))))))))))))...(((((...........))))).... ( -38.30) >DroSec_CAF1 84036 111 - 1 AAAUUUGAAGGUGUU---------GCUGGUGUUUUUGGCGAUGUUGCUGCUGUCGCGGCGACAACAAUGAAGAUGACAGCAACAGCAACAUCGCAGUUUCUGGCUUAUUUUAAGGCAACA .........(.((((---------((((..(((((((..(.(((((((((....))))))))).)..)))))))..)))))))).)................((((......)))).... ( -36.40) >DroSim_CAF1 76958 111 - 1 AAAUUUGAAGGUGUU---------GCUGGGGUUUUUGGCCAUGUUGCUGCUGUCGCGGCGACAACAAUGAAGAUGACAGCAACAGCAACAUCACAGUUUCUGGCUUAUUUUGAGGCAACA .........(.((((---------((((..(((((((....(((((((((....)))))))))....)))))))..)))))))).).......(((...)))((((......)))).... ( -37.30) >DroYak_CAF1 81030 119 - 1 AAAUCUGGGAGUGUUGCUGGUGUUUAUGAUGUUGGUGGCCAUGUUGCUGCUGUCGCGGCGACAACAAUGAAGAUGACAGCAACAGCAACACCACAGUUUCUGGCUUGUUUUG-GGCAACA ....(((.(.(((((((((.((((..(.((.((.((.....(((((((((....)))))))))...)).)).)).).)))).)))))))))).)))......((((.....)-))).... ( -38.10) >consensus AAAUUUGAAGGUGUU_________GCUGGUGUUUUUGGCCAUGUUGCUGCUGUCGCGGCGACAACAAUGAAGAUGACAGCAACAGCAACAUCACAGUUUCUGGCUUAUUUUGAGGCAACA .........((((((((((.....((((..(((((((....(((((((((....)))))))))....)))))))..))))..))))))))))..........((((......)))).... (-28.79 = -32.23 + 3.44)



| Location | 14,948,383 – 14,948,503 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.47 |
| Mean single sequence MFE | -36.78 |
| Consensus MFE | -28.27 |
| Energy contribution | -29.21 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14948383 120 - 27905053 AAGUGUGUCACUUGAACCAAAACACCAAGUUUAGGUGCUGAAAUUUGAAGGUGUUGCUGAUGUUGCUGGUGUUUUUGGCCAUGCUGCUGCUGUCGCGGCGACAACAAUGAAGAUGACAGC .....((((((((.(..((.((((((...(((((((......)))))))))))))..)).((((((((((.......)))).)).(((((....)))))))))....).))).))))).. ( -32.30) >DroSec_CAF1 84076 111 - 1 AAGUGUGUCACUUGAACCAAAACACCAAGUUUAGGCGCUCAAAUUUGAAGGUGUU---------GCUGGUGUUUUUGGCGAUGUUGCUGCUGUCGCGGCGACAACAAUGAAGAUGACAGC .....((((((((.(.(((((((((((.((...((((((((....)))..)))))---------))))))).)))))).(.(((((((((....))))))))).)..).))).))))).. ( -37.60) >DroSim_CAF1 76998 111 - 1 AAGUGUGUCACUUGAACCAAAACACCAAGUUUAGGCGCACAAAUUUGAAGGUGUU---------GCUGGGGUUUUUGGCCAUGUUGCUGCUGUCGCGGCGACAACAAUGAAGAUGACAGC ..((((((((((((...........)))))...)))))))...............---------((((..(((((((....(((((((((....)))))))))....)))))))..)))) ( -36.40) >DroYak_CAF1 81069 120 - 1 AAGUGUGUCACUUGAACCAAAACACCAAGUUUAGGCGCUCAAAUCUGGGAGUGUUGCUGGUGUUUAUGAUGUUGGUGGCCAUGUUGCUGCUGUCGCGGCGACAACAAUGAAGAUGACAGC .....((((((((.(((((((((((((.((...(((((((........)))))))))))))))).......))))).....(((((((((....)))))))))....).))).))))).. ( -40.81) >consensus AAGUGUGUCACUUGAACCAAAACACCAAGUUUAGGCGCUCAAAUUUGAAGGUGUU_________GCUGGUGUUUUUGGCCAUGUUGCUGCUGUCGCGGCGACAACAAUGAAGAUGACAGC .....((((((((.(.(((((((((((......((((((..........))))))...........))))))).))))...(((((((((....)))))))))....).))).))))).. (-28.27 = -29.21 + 0.94)



| Location | 14,948,423 – 14,948,543 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.09 |
| Mean single sequence MFE | -33.75 |
| Consensus MFE | -23.84 |
| Energy contribution | -23.52 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.581030 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14948423 120 - 27905053 AGCCAUUAACUUCUUCAUAUGGCUGUGAGCAUCUCUACGCAAGUGUGUCACUUGAACCAAAACACCAAGUUUAGGUGCUGAAAUUUGAAGGUGUUGCUGAUGUUGCUGGUGUUUUUGGCC .((((..(((....(((..((((...(((...)))(((....))).))))..)))((((.((((((...(((((((......)))))))))))))((.......)))))))))..)))). ( -31.80) >DroSec_CAF1 84116 109 - 1 AGCCAUUAACUUCUUCAUAUGGCUGUGAGCAUC--UACGCAAGUGUGUCACUUGAACCAAAACACCAAGUUUAGGCGCUCAAAUUUGAAGGUGUU---------GCUGGUGUUUUUGGCG .((((..(((....(((..((((((....))..--(((....))).))))..)))((((.((((((..((....))..(((....))).))))))---------..)))))))..)))). ( -30.80) >DroSim_CAF1 77038 109 - 1 AGCCAUUAACUUCUUCAUAUGGCUGUGAGCAUC--UACGCAAGUGUGUCACUUGAACCAAAACACCAAGUUUAGGCGCACAAAUUUGAAGGUGUU---------GCUGGGGUUUUUGGCC .((((..((((((.(((((....)))))(((((--(...(((((((((((((((...........)))))...)))))))....))).)))))).---------...))))))..)))). ( -31.70) >DroYak_CAF1 81109 118 - 1 AGCCAUUAACUUCUUCAUAUAGCUGUGAGCAUC--UGCGCAAGUGUGUCACUUGAACCAAAACACCAAGUUUAGGCGCUCAAAUCUGGGAGUGUUGCUGGUGUUUAUGAUGUUGGUGGCC .(((((((((....((((......((((.(((.--.((....)))))))))........((((((((.((...(((((((........))))))))))))))))))))).))))))))). ( -40.70) >consensus AGCCAUUAACUUCUUCAUAUGGCUGUGAGCAUC__UACGCAAGUGUGUCACUUGAACCAAAACACCAAGUUUAGGCGCUCAAAUUUGAAGGUGUU_________GCUGGUGUUUUUGGCC .((((..(((.((.(((((....)))))(((((...((....))((((((((((...........)))))...)))))...........))))).............)).)))..)))). (-23.84 = -23.52 + -0.31)

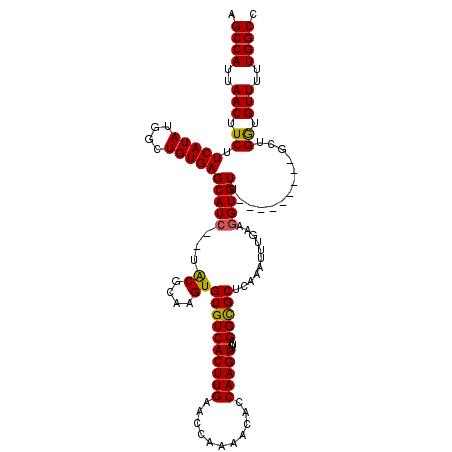

| Location | 14,948,503 – 14,948,623 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.40 |
| Mean single sequence MFE | -23.59 |
| Consensus MFE | -19.56 |
| Energy contribution | -20.31 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14948503 120 - 27905053 UCUAGUGAACCACUUUACACAACUUCCAGCGGCAACAUAGCCAUUCCGCCGCUAACAAUGUAACUAUUUUGAGCUCUUUAAGCCAUUAACUUCUUCAUAUGGCUGUGAGCAUCUCUACGC ..(((.((......(((((........((((((..............)))))).....))))).........((((....((((((............))))))..)))).)).)))... ( -26.46) >DroSec_CAF1 84187 118 - 1 UCUAGUGAACCACUUUACACAACUCCCAGCGGCAACAUCACCAUCCCGCCGCUAACAAUGUAACUAUUUUGAGCUCUUUAAGCCAUUAACUUCUUCAUAUGGCUGUGAGCAUC--UACGC ....(((((....))))).........((((((..............))))))...................((((....((((((............))))))..))))...--..... ( -25.64) >DroSim_CAF1 77109 118 - 1 UCUAGUGAACCACUUUACACAACUCUCAGCGGCAACAUCGCCAUCCCGCCGCUAACAAUGUAACUAUUUUGAGCUCUUUAAGCCAUUAACUUCUUCAUAUGGCUGUGAGCAUC--UACGC ....(((((....))))).........((((((..............))))))...................((((....((((((............))))))..))))...--..... ( -25.64) >DroYak_CAF1 81189 118 - 1 UCUACUGAACCACUUUACACUACUCCCAGCGGCAACAUCACCAUCCCACCGCUAACAAUGUAACUAUUUUGAGGUCUUUAAGCCAUUAACUUCUUCAUAUAGCUGUGAGCAUC--UGCGC ..........(((.(((((........(((((................))))).....)))))((((..(((((...((((....))))...))))).))))..))).((...--.)).. ( -16.61) >consensus UCUAGUGAACCACUUUACACAACUCCCAGCGGCAACAUCACCAUCCCGCCGCUAACAAUGUAACUAUUUUGAGCUCUUUAAGCCAUUAACUUCUUCAUAUGGCUGUGAGCAUC__UACGC ..............(((((........((((((..............)))))).....))))).........((((....((((((............))))))..)))).......... (-19.56 = -20.31 + 0.75)

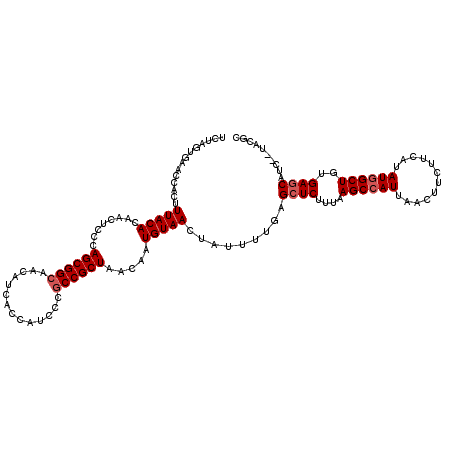
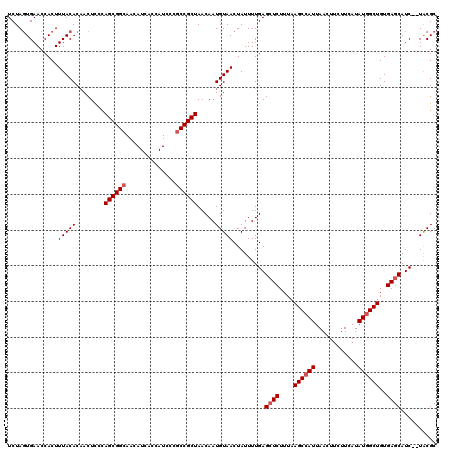
| Location | 14,948,543 – 14,948,654 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 94.89 |
| Mean single sequence MFE | -31.85 |
| Consensus MFE | -29.92 |
| Energy contribution | -30.05 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632648 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14948543 111 + 27905053 UAAAGAGCUCAAAAUAGUUACAUUGUUAGCGGCGGAAUGGCUAUGUUGCCGCUGGAAGUUGUGUAAAGUGGUUCACUAGAAACGCCCCAAAAUUGUCGGAGGCAUUCUAAG ....((((.((......((((((..((((((((((.((....)).)))))))))).....))))))..))))))..(((((..(((((.........)).))).))))).. ( -30.80) >DroSec_CAF1 84225 111 + 1 UAAAGAGCUCAAAAUAGUUACAUUGUUAGCGGCGGGAUGGUGAUGUUGCCGCUGGGAGUUGUGUAAAGUGGUUCACUAGAAACGCCCCAAAAUUGUCGGAGGCAUUCUAAG ....((((.((......(((((((.((((((((((.((....)).)))))))))).)...))))))..))))))..(((((..(((((.........)).))).))))).. ( -32.90) >DroSim_CAF1 77147 111 + 1 UAAAGAGCUCAAAAUAGUUACAUUGUUAGCGGCGGGAUGGCGAUGUUGCCGCUGAGAGUUGUGUAAAGUGGUUCACUAGAAACGCCCCAAAAUUGUCGGAGGCAUUCUAAG ....((((.((......(((((((.((((((((((.((....)).)))))))))).)...))))))..))))))..(((((..(((((.........)).))).))))).. ( -33.70) >DroYak_CAF1 81227 111 + 1 UAAAGACCUCAAAAUAGUUACAUUGUUAGCGGUGGGAUGGUGAUGUUGCCGCUGGGAGUAGUGUAAAGUGGUUCAGUAGAAACGCCCCAAAAUUGUUGGAGGCAUUCUUAG ....((((.(.......(((((((((((((((..(.((....)).)..)))))))...)))))))).).))))....((((..(((((((.....)))).))).))))... ( -30.00) >consensus UAAAGAGCUCAAAAUAGUUACAUUGUUAGCGGCGGGAUGGCGAUGUUGCCGCUGGGAGUUGUGUAAAGUGGUUCACUAGAAACGCCCCAAAAUUGUCGGAGGCAUUCUAAG ....((((.((......((((((..((((((((((.((....)).)))))))))).....))))))..))))))..(((((..(((((.........)).))).))))).. (-29.92 = -30.05 + 0.13)


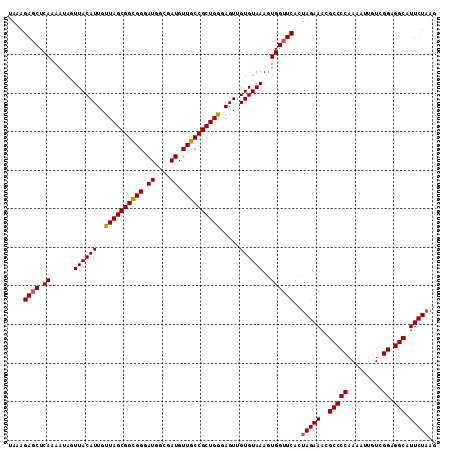
| Location | 14,948,543 – 14,948,654 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 94.89 |
| Mean single sequence MFE | -23.73 |
| Consensus MFE | -21.73 |
| Energy contribution | -22.29 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14948543 111 - 27905053 CUUAGAAUGCCUCCGACAAUUUUGGGGCGUUUCUAGUGAACCACUUUACACAACUUCCAGCGGCAACAUAGCCAUUCCGCCGCUAACAAUGUAACUAUUUUGAGCUCUUUA (((((((((((.((((.....)))))))(((....(((((....))))).........((((((..............))))))))).........))))))))....... ( -24.94) >DroSec_CAF1 84225 111 - 1 CUUAGAAUGCCUCCGACAAUUUUGGGGCGUUUCUAGUGAACCACUUUACACAACUCCCAGCGGCAACAUCACCAUCCCGCCGCUAACAAUGUAACUAUUUUGAGCUCUUUA (((((((((((.((((.....)))))))(((....(((((....))))).........((((((..............))))))))).........))))))))....... ( -24.94) >DroSim_CAF1 77147 111 - 1 CUUAGAAUGCCUCCGACAAUUUUGGGGCGUUUCUAGUGAACCACUUUACACAACUCUCAGCGGCAACAUCGCCAUCCCGCCGCUAACAAUGUAACUAUUUUGAGCUCUUUA (((((((((((.((((.....)))))))(((....(((((....))))).........((((((..............))))))))).........))))))))....... ( -24.94) >DroYak_CAF1 81227 111 - 1 CUAAGAAUGCCUCCAACAAUUUUGGGGCGUUUCUACUGAACCACUUUACACUACUCCCAGCGGCAACAUCACCAUCCCACCGCUAACAAUGUAACUAUUUUGAGGUCUUUA .((..((((((.((((.....))))))))))..))....(((................(((((................)))))..(((..........))).)))..... ( -20.09) >consensus CUUAGAAUGCCUCCGACAAUUUUGGGGCGUUUCUAGUGAACCACUUUACACAACUCCCAGCGGCAACAUCACCAUCCCGCCGCUAACAAUGUAACUAUUUUGAGCUCUUUA (((((((((((.((((.....)))))))(((....(((((....))))).........((((((..............))))))))).........))))))))....... (-21.73 = -22.29 + 0.56)

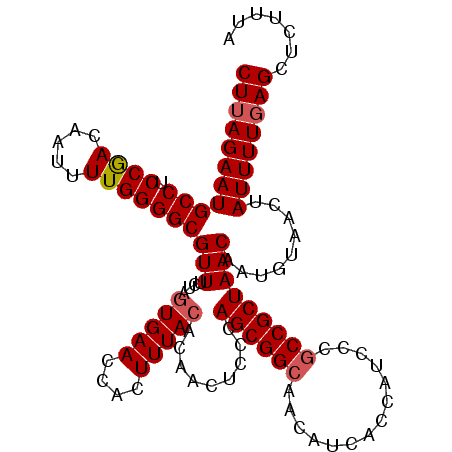
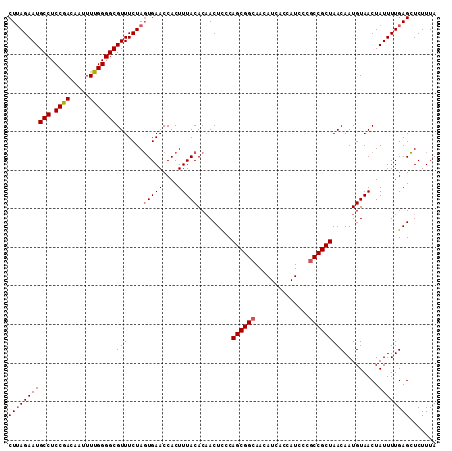
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:23 2006