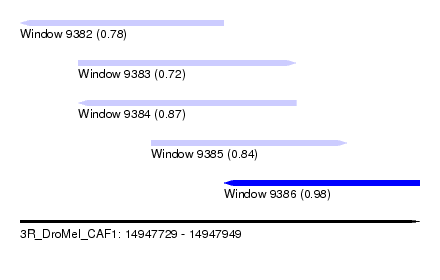
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,947,729 – 14,947,949 |
| Length | 220 |
| Max. P | 0.979838 |

| Location | 14,947,729 – 14,947,841 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.26 |
| Mean single sequence MFE | -28.05 |
| Consensus MFE | -18.86 |
| Energy contribution | -18.93 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14947729 112 - 27905053 GAGCGAACAUAAAAAAAAAAAAAACGGGCAAAAGGCGAUGAGAGUUGUGCCCAUUCUCAUGGAAAGCAAGUGUAUCGCCAGAUCACAUAUA-----UGUCUGCA---UGUUGGCCAACUA ..........................(((....(((((((..(.((((..((((....))))...)))).).)))))))......((.(((-----((....))---))))))))..... ( -26.90) >DroSec_CAF1 83413 119 - 1 GAACGAACAUAUAAAAA-AAAAAACAGGCAAACGGCGAUGAGAGUUGUGCCCAUUCUCAUGGAAAGCAAGUGGAUCGCCAGAUCACAUAUAUGGUAUGUAUGUAUGAUGUUGGCCAACUA .................-........(((.(((((((((...(.((((..((((....))))...)))).)..))))))..((((.((((((.....)))))).))))))).)))..... ( -29.40) >DroSim_CAF1 76337 114 - 1 GAGCGAACAUAUAAAAA-AAAAAACAGGCAAAAGGCGAUGAGAGUUGUGCCCAUUCUCAUGGAAAGCAAGUGGAUCGCCAGAUCACAUAUA-----UGUAUGUAUGAUGUUGGCCAACUA (.((.(((((.......-...............((((((...(.((((..((((....))))...)))).)..))))))......((((((-----....))))))))))).)))..... ( -26.90) >DroYak_CAF1 80424 97 - 1 CAGCGAACAUA-------AAAAAACGGGCAAAAGGCGAUGAGAGCUGUGCCCAUUCUCAUGGAAAGCAAGUGGAUCGGCAGAUCGCAUAUA----------------UGUUGGCCAACUA ..((.((((((-------.......(((((...(((.......))).))))).................(((((((....)))).))).))----------------)))).))...... ( -29.00) >consensus GAGCGAACAUAUAAAAA_AAAAAACAGGCAAAAGGCGAUGAGAGUUGUGCCCAUUCUCAUGGAAAGCAAGUGGAUCGCCAGAUCACAUAUA_____UGUAUGUA___UGUUGGCCAACUA ..((.((((..................((......(.(((((((.((....))))))))).)...))..(((.(((....)))))).....................)))).))...... (-18.86 = -18.93 + 0.06)



| Location | 14,947,761 – 14,947,881 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.47 |
| Mean single sequence MFE | -31.06 |
| Consensus MFE | -26.20 |
| Energy contribution | -26.02 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14947761 120 + 27905053 UGGCGAUACACUUGCUUUCCAUGAGAAUGGGCACAACUCUCAUCGCCUUUUGCCCGUUUUUUUUUUUUUUAUGUUCGCUCAACUUGUGUGUGGCCAUAAAUAAAUGGCCAAUCGAUUCCG .(((((.(((............(((((((((((.((..(.....)..)).)))))))))))..........))))))))....(((....(((((((......)))))))..)))..... ( -30.25) >DroSec_CAF1 83453 119 + 1 UGGCGAUCCACUUGCUUUCCAUGAGAAUGGGCACAACUCUCAUCGCCGUUUGCCUGUUUUUU-UUUUUAUAUGUUCGUUCAGCUUGUGUGUGGCCAUAAAUAAAUGGCCAAUCGAUUCCG .((.(((((((..((.......(((((((((((.(((..........)))))))))))))).-........((......))))..)))..(((((((......)))))))...)))))). ( -30.50) >DroSim_CAF1 76372 119 + 1 UGGCGAUCCACUUGCUUUCCAUGAGAAUGGGCACAACUCUCAUCGCCUUUUGCCUGUUUUUU-UUUUUAUAUGUUCGCUCAGCUUGUGUGUGGCCAUAAAUAAAUGGCCAAUCGAUUCCG .((.(((((((..((.......(((((((((((.((..(.....)..)).))))))))))).-........((......))))..)))..(((((((......)))))))...)))))). ( -28.80) >DroYak_CAF1 80448 113 + 1 UGCCGAUCCACUUGCUUUCCAUGAGAAUGGGCACAGCUCUCAUCGCCUUUUGCCCGUUUUUU-------UAUGUUCGCUGAGUUAGUGUGUGGCCAUAAAUAAAUGGCCAAUCGAUUCCG ...((((.((((.((((..((((((((((((((..((.......))....)))))))))..)-------))))......)))).))))..(((((((......)))))))))))...... ( -34.70) >consensus UGGCGAUCCACUUGCUUUCCAUGAGAAUGGGCACAACUCUCAUCGCCUUUUGCCCGUUUUUU_UUUUUAUAUGUUCGCUCAGCUUGUGUGUGGCCAUAAAUAAAUGGCCAAUCGAUUCCG ...((((.(((..(((......(((((((((((.................)))))))))))...................)))..)))..(((((((......)))))))))))...... (-26.20 = -26.02 + -0.19)

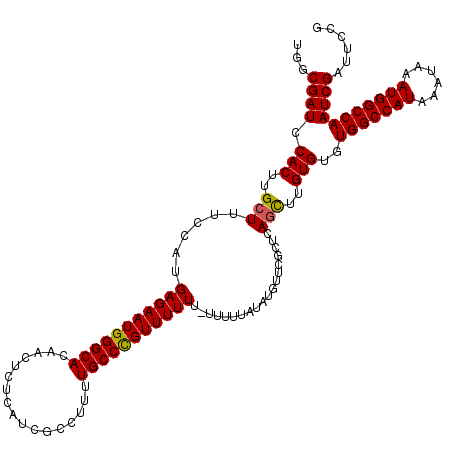
| Location | 14,947,761 – 14,947,881 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.47 |
| Mean single sequence MFE | -31.45 |
| Consensus MFE | -26.12 |
| Energy contribution | -27.00 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14947761 120 - 27905053 CGGAAUCGAUUGGCCAUUUAUUUAUGGCCACACACAAGUUGAGCGAACAUAAAAAAAAAAAAAACGGGCAAAAGGCGAUGAGAGUUGUGCCCAUUCUCAUGGAAAGCAAGUGUAUCGCCA .....((((((((((((......))))))).......)))))((((((((...............((((.....(((((....))))))))).(((.....))).....)))).)))).. ( -32.01) >DroSec_CAF1 83453 119 - 1 CGGAAUCGAUUGGCCAUUUAUUUAUGGCCACACACAAGCUGAACGAACAUAUAAAAA-AAAAAACAGGCAAACGGCGAUGAGAGUUGUGCCCAUUCUCAUGGAAAGCAAGUGGAUCGCCA .((...(((((((((((......))))))...(((..(((...(.............-.........((.....)).((((((((.......)))))))))...)))..)))))))))). ( -30.30) >DroSim_CAF1 76372 119 - 1 CGGAAUCGAUUGGCCAUUUAUUUAUGGCCACACACAAGCUGAGCGAACAUAUAAAAA-AAAAAACAGGCAAAAGGCGAUGAGAGUUGUGCCCAUUCUCAUGGAAAGCAAGUGGAUCGCCA .((...(((((((((((......))))))...(((..(((...(.............-.........((.....)).((((((((.......)))))))).)..)))..)))))))))). ( -30.00) >DroYak_CAF1 80448 113 - 1 CGGAAUCGAUUGGCCAUUUAUUUAUGGCCACACACUAACUCAGCGAACAUA-------AAAAAACGGGCAAAAGGCGAUGAGAGCUGUGCCCAUUCUCAUGGAAAGCAAGUGGAUCGGCA .....((((((((((((......))))))...((((...............-------.......(((((...(((.......))).))))).(((.....)))....)))))))))).. ( -33.50) >consensus CGGAAUCGAUUGGCCAUUUAUUUAUGGCCACACACAAGCUGAGCGAACAUAUAAAAA_AAAAAACAGGCAAAAGGCGAUGAGAGUUGUGCCCAUUCUCAUGGAAAGCAAGUGGAUCGCCA .((...(((((((((((......))))))...(((..(((.........................(((((...(((.......))).))))).(((.....))))))..)))))))))). (-26.12 = -27.00 + 0.88)

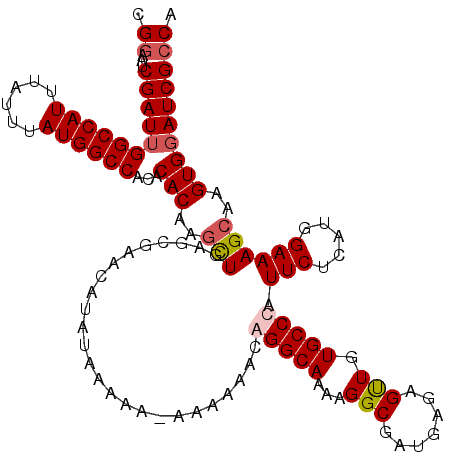
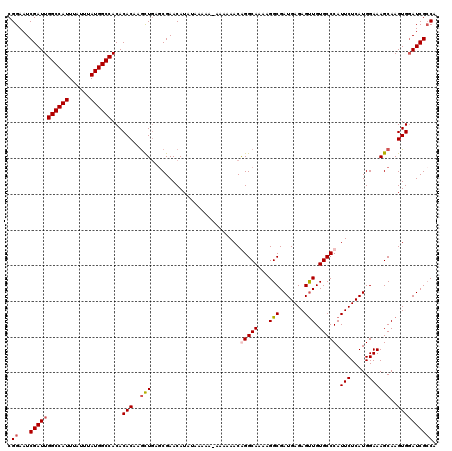
| Location | 14,947,801 – 14,947,909 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.70 |
| Mean single sequence MFE | -30.20 |
| Consensus MFE | -21.69 |
| Energy contribution | -23.25 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14947801 108 + 27905053 CAUCGCCUUUUGCCCGUUUUUUUUUUUUUUAUGUUCGCUCAACUUGUGUGUGGCCAUAAAUAAAUGGCCAAUCGAUUCCGCAACCGAAUGCAAAUGAGUGAAAGAAAA------------ ..........................((((...((((((((..((((((.(((((((......))))))).(((..........))))))))).))))))))..))))------------ ( -24.80) >DroSec_CAF1 83493 119 + 1 CAUCGCCGUUUGCCUGUUUUUU-UUUUUAUAUGUUCGUUCAGCUUGUGUGUGGCCAUAAAUAAAUGGCCAAUCGAUUCCGCAACCGAAUGCAAAUGAGUGAAAGAAAAAAUGCAAACAAU .......((((((..(((((((-(((((((.(((((((((.(.(((((..(((((((......)))))))........)))))).)))))..)))).))))))))))))))))))))... ( -34.80) >DroSim_CAF1 76412 119 + 1 CAUCGCCUUUUGCCUGUUUUUU-UUUUUAUAUGUUCGCUCAGCUUGUGUGUGGCCAUAAAUAAAUGGCCAAUCGAUUCCGCAACCGAAUGCAAAUGAGUGAAAGAAAAAAUGCAAACAAU ........(((((..(((((((-(((........(((((((..((((((.(((((((......))))))).(((..........))))))))).)))))))))))))))))))))).... ( -31.80) >DroYak_CAF1 80488 113 + 1 CAUCGCCUUUUGCCCGUUUUUU-------UAUGUUCGCUGAGUUAGUGUGUGGCCAUAAAUAAAUGGCCAAUCGAUUCCGCAACCGAAUGCAAAUGAGUGAAAGAAAAAAUGCAAACAAU ........(((((..(((((((-------(...((((((((((..((...(((((((......)))))))))..)))).(((......))).....)))))).))))))))))))).... ( -29.40) >consensus CAUCGCCUUUUGCCCGUUUUUU_UUUUUAUAUGUUCGCUCAGCUUGUGUGUGGCCAUAAAUAAAUGGCCAAUCGAUUCCGCAACCGAAUGCAAAUGAGUGAAAGAAAAAAUGCAAACAAU ........(((((...((((((...........((((((((...(((((.(((((((......))))))).(((..........))))))))..))))))))))))))...))))).... (-21.69 = -23.25 + 1.56)

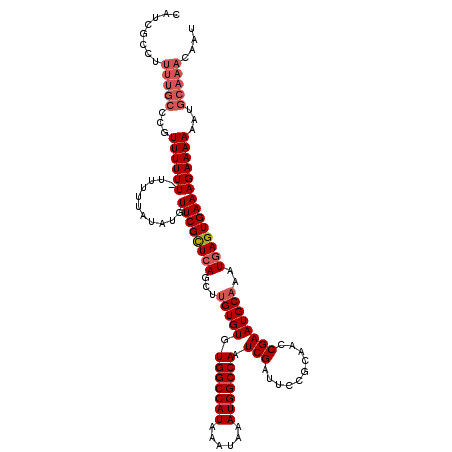

| Location | 14,947,841 – 14,947,949 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.63 |
| Mean single sequence MFE | -24.53 |
| Consensus MFE | -21.02 |
| Energy contribution | -20.95 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979838 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14947841 108 - 27905053 UGUCUAUUUCUGCCACGGAUUUUUUUUGUUUUCAUGUUAA------------UUUUCUUUCACUCAUUUGCAUUCGGUUGCGGAAUCGAUUGGCCAUUUAUUUAUGGCCACACACAAGUU .(((...((((((.((.((.......(((......((.((------------......)).))......))).)).)).))))))..)))(((((((......))))))).......... ( -21.70) >DroSec_CAF1 83532 119 - 1 UGUCUAUUUCUGCCACGGAUUUU-UUUGUUUUCUUGUUUAAUUGUUUGCAUUUUUUCUUUCACUCAUUUGCAUUCGGUUGCGGAAUCGAUUGGCCAUUUAUUUAUGGCCACACACAAGCU .(((...((((((.(((((....-)))))..........(((((..((((..................))))..)))))))))))..)))(((((((......))))))).......... ( -25.57) >DroSim_CAF1 76451 119 - 1 UGUCUAUUUCUGCCACGGAUUUU-UUUGUUUUCUUGUUUAAUUGUUUGCAUUUUUUCUUUCACUCAUUUGCAUUCGGUUGCGGAAUCGAUUGGCCAUUUAUUUAUGGCCACACACAAGCU .(((...((((((.(((((....-)))))..........(((((..((((..................))))..)))))))))))..)))(((((((......))))))).......... ( -25.57) >DroYak_CAF1 80521 118 - 1 UAUCUAUUUCUGCCACGGAUUUU-UUUGUUUUCAUGUU-AAUUGUUUGCAUUUUUUCUUUCACUCAUUUGCAUUCGGUUGCGGAAUCGAUUGGCCAUUUAUUUAUGGCCACACACUAACU .(((...((((((.(((((....-))))).........-(((((..((((..................))))..)))))))))))..)))(((((((......))))))).......... ( -25.27) >consensus UGUCUAUUUCUGCCACGGAUUUU_UUUGUUUUCAUGUUUAAUUGUUUGCAUUUUUUCUUUCACUCAUUUGCAUUCGGUUGCGGAAUCGAUUGGCCAUUUAUUUAUGGCCACACACAAGCU .(((...((((((.((.((.......((..............((....))..............)).......)).)).))))))..)))(((((((......))))))).......... (-21.02 = -20.95 + -0.06)


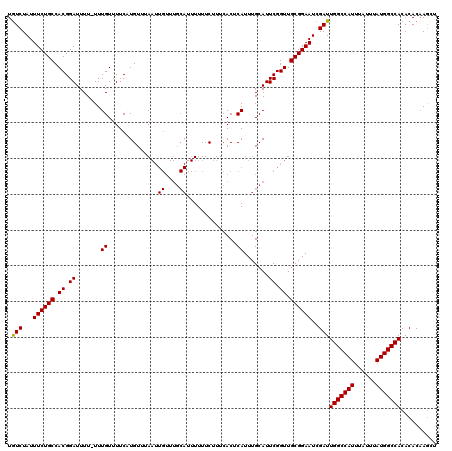
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:13 2006