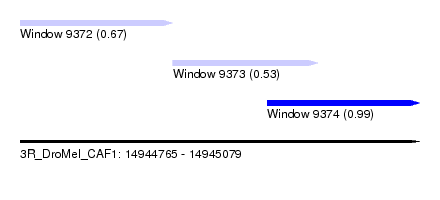
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,944,765 – 14,945,079 |
| Length | 314 |
| Max. P | 0.992519 |

| Location | 14,944,765 – 14,944,885 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.80 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -25.76 |
| Energy contribution | -26.26 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14944765 120 + 27905053 CCCAACCAACGCCACCAACUUCUCCCUUCUUUUUUUUUUAUUUUCUUUGCAACACUAAGAGGCAGGAAGAUGUGGGCGCGCUCACAGGCAUAAAUUCACAGGCUUUGGUUGAUUAAGGCA ..(((((((.(((.....(((((..((((((.........................))))))..))))).((((((....))))))..............))).)))))))......... ( -32.31) >DroSec_CAF1 80582 115 + 1 CC-AACCAACGCCACCAACUUCCCCCUUCUUUUUUUC----UUUCUUUGCAACACUAAGAGGCAGGAAGAUGUGGGCGCGCUCACAGGCAUAAAUUCACAGGCUUUGGUUGAUUAAGGCA .(-((((((.(((.....(((((..((((((......----...............))))))..))))).((((((....))))))..............))).)))))))......... ( -34.10) >DroSim_CAF1 73415 115 + 1 CC-CACCAACGCCACCAACUUCCCCCUUCUUUUUUUC----UUUCUUUGCAACACUAAGAGGCAGGAAGAUGUGGGCGCGCUCACAGGCAUAAAUUCACAGGCUUUGGUUGAUUAAGGCA .(-.(((((.(((.....(((((..((((((......----...............))))))..))))).((((((....))))))..............))).))))).)......... ( -30.00) >DroYak_CAF1 77528 107 + 1 CC-AGCCACCGCCACCCACUUCCCCCCUUAUUU------------CUUGCAACACCAAGAGGCAGUAAGAUGUGGGCGCGCUCACAGGCAUAAAUUUACAGGCUUUGCUUGAUUAAGGAG ..-.....(((((.............(((((((------------((((......)))))...)))))).((((((....))))))))).........(((((...))))).....)).. ( -26.00) >consensus CC_AACCAACGCCACCAACUUCCCCCUUCUUUUUUUC____UUUCUUUGCAACACUAAGAGGCAGGAAGAUGUGGGCGCGCUCACAGGCAUAAAUUCACAGGCUUUGGUUGAUUAAGGCA ...((((((.(((.....(((((..((((((.........................))))))..))))).((((((....))))))..............))).)))))).......... (-25.76 = -26.26 + 0.50)

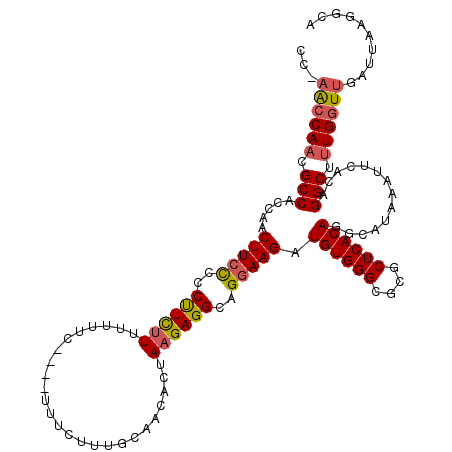
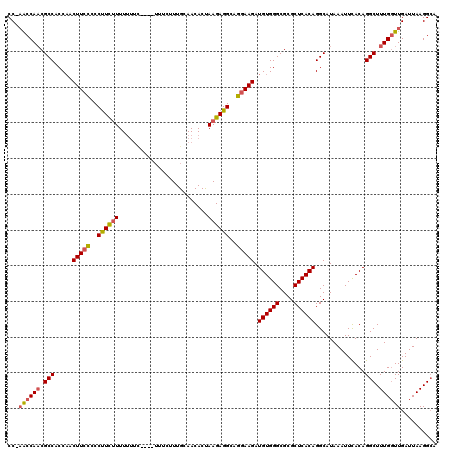
| Location | 14,944,885 – 14,944,999 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.77 |
| Mean single sequence MFE | -23.35 |
| Consensus MFE | -18.59 |
| Energy contribution | -18.40 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14944885 114 + 27905053 GCUCCCCCGCAGCUUUUGC------UGCUCCCCCCUCCGCAUUUUCCUGGAAUAUCCUUGGCGAAAAUGAACAUAAAACAUUUCUCCGUAAUCAACUGGGCUGCUAAAAAUAAACCCAAA ((..(((.(((((....))------)))...........(((((((((((.....))..)).)))))))............................)))..))................ ( -23.70) >DroSec_CAF1 80697 116 + 1 GCUCCCCCGCAGCUCUGGC---UGCUGCUC-CUCCUUCGCAUUUUCCUGGAAUAUCCUUGGCGAAAAUGAACAUAAAACAUUUCUCCGUAAUCAACUGGGCUGCUAAAAAUAAACCCAAA ((..(((.(((((....))---))).....-........(((((((((((.....))..)).)))))))............................)))..))................ ( -23.50) >DroSim_CAF1 73530 119 + 1 GCUCCCCCGCAGCUCUUGCUCUUGCUGCUC-CUCCUCCGCAUUUUCCUGGAAUAUCCUUGGCGAAAAUGAACAUAAAACAUUUCUCCGUAAUCAACUGGGCUGCUAAAAAUAAACCCAAA ((..(((.(((((..........)))))..-........(((((((((((.....))..)).)))))))............................)))..))................ ( -22.80) >DroYak_CAF1 77635 111 + 1 GCUC--CCGCAGCUCUUG------CUGCUC-CUCCUCCGCAUUUUCCUGGAAUAUCCUUGGCGAAAAUGAACAUAAAACAUUUCUCCGUAAUCAACUGGGCUGCUAAAAAUAAACCCAAA ((.(--(((((((....)------))))..-........(((((((((((.....))..)).)))))))............................)))..))................ ( -23.40) >consensus GCUCCCCCGCAGCUCUUGC_____CUGCUC_CUCCUCCGCAUUUUCCUGGAAUAUCCUUGGCGAAAAUGAACAUAAAACAUUUCUCCGUAAUCAACUGGGCUGCUAAAAAUAAACCCAAA ........(((((((........................(((((((((((.....))..)).)))))))........((........))........)))))))................ (-18.59 = -18.40 + -0.19)


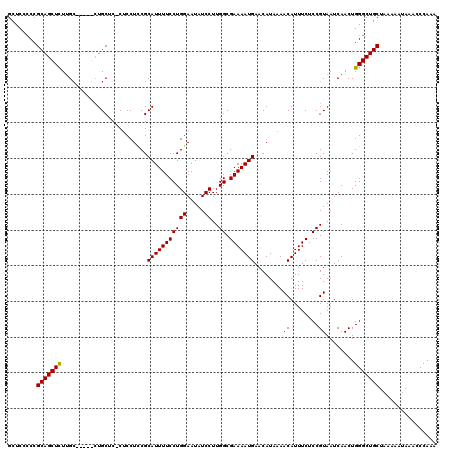
| Location | 14,944,959 – 14,945,079 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.10 |
| Mean single sequence MFE | -42.00 |
| Consensus MFE | -40.06 |
| Energy contribution | -40.25 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.75 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14944959 120 + 27905053 UUUCUCCGUAAUCAACUGGGCUGCUAAAAAUAAACCCAAACUUGGCCUGCGAGGCCUUGUGGGCUAAUGCUGUAAGGCCUUUUGGUUUAGCAGCUUAGCAAGUAUGGCAACUUCAAUUAA .....(((((.....(((((((((((((.......(((....)))((...(((((((((..(.(....))..)))))))))..)))))))))))))))....)))))............. ( -42.30) >DroSec_CAF1 80773 118 + 1 UUUCUCCGUAAUCAACUGGGCUGCUAAAAAUAAACCCAAACUUGGCCUGCGGGGCCUU--GGGCUAAUGCUGUAAGGCCUUUUGGUUUAGCGGCUUAGCAAGUAUGGCAACUUCAAUUAA .....(((((.....(((((((((((((.......(((((...(((((((((((((..--.))))....)))).))))).))))))))))))))))))....)))))............. ( -40.31) >DroSim_CAF1 73609 118 + 1 UUUCUCCGUAAUCAACUGGGCUGCUAAAAAUAAACCCAAACUUGGCCUGCGGGGCCUU--GGGCUAAUGCUCUAAGGCCUUUUGGUUUAGCGGCUUAGCAAGUAUGGCAACUUCAAUUAA .....(((((.....(((((((((((((.......(((....)))((...((((((((--((((....)).))))))))))..)))))))))))))))....)))))............. ( -43.10) >DroYak_CAF1 77706 116 + 1 UUUCUCCGUAAUCAACUGGGCUGCUAAAAAUAAACCCAAACUCGGCCUG--GGGCCUU--GGGCUAACGCCAAAAGGCCUUUUGGUUUAGCGGCUUAGCAAGUAUGGCAACUUCAAUUAA .....(((((.....(((((((((((((.......((......))((.(--(((((((--.(((....)))..))))))))..)))))))))))))))....)))))............. ( -42.30) >consensus UUUCUCCGUAAUCAACUGGGCUGCUAAAAAUAAACCCAAACUUGGCCUGCGGGGCCUU__GGGCUAAUGCUGUAAGGCCUUUUGGUUUAGCGGCUUAGCAAGUAUGGCAACUUCAAUUAA .....(((((.....(((((((((((((.......((......))((...((((((((...(((....)))..))))))))..)))))))))))))))....)))))............. (-40.06 = -40.25 + 0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:01 2006