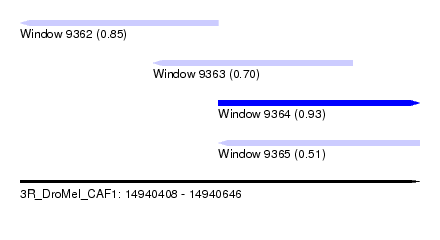
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,940,408 – 14,940,646 |
| Length | 238 |
| Max. P | 0.928012 |

| Location | 14,940,408 – 14,940,526 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.55 |
| Mean single sequence MFE | -27.10 |
| Consensus MFE | -19.29 |
| Energy contribution | -20.85 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14940408 118 - 27905053 ACAA-AAGUAUAGUGUAUAUAUUUAUGUAUAAGCCGAAUUCCAGAAAACAAAUUCUCCGUCAAAUGGAUGCGAUGUUCGAGGGGAAGGGGAGG-GGGAUUCGAGCGAUGAAUGCGGCAAC (((.-(((((((.....))))))).)))....((((((((((......(...(((((((((.....))).((.....)).))))))..)....-)))))))..(((.....))))))... ( -30.00) >DroSec_CAF1 76231 108 - 1 ACAAAAAGUAUAG--UAUAUAUUUAUGUAAAAGCCGAAUUCCAGAAAACAAAUUCUCCGUCAAAUGGACGGGAUG----------AGGGAAGGGGGGAUUCGAGCGAUGAGUGCGGCAAC (((..(((((((.--..))))))).)))....((((((((((......(....((.(((((.....)))))))..----------..)......)))))))..(((.....))))))... ( -27.20) >DroSim_CAF1 69052 107 - 1 ACAAAAAGUAUAG--UAUAUAUUUAUGUAUAAGCCGAAUUCCAGAAAACAAAUUCUCCGUCAAAUGGAUGGGAUG----------AGGGGAUG-GUGAUUCGAGCGAUGAGUGCGGCAAC .......((((.(--(((((....))))))..(((((((((((.....(....((.(((((.....)))))))..----------..)...))-).)))))).)).....))))...... ( -23.50) >DroYak_CAF1 73298 94 - 1 ACAAAAAGUAUAA--UAUAU--------AUAAGCCGAAUUCCAGAAAACAAAUUCUCCGUCAAAUGGAUG-GAUG----------UU----GG-GGGAUUCGAGCGCUGAGUGCGGCAAC .............--.....--------....((((((((((....((((.....((((((.....))))-))))----------))----..-)))))))).))((((....))))... ( -27.70) >consensus ACAAAAAGUAUAG__UAUAUAUUUAUGUAUAAGCCGAAUUCCAGAAAACAAAUUCUCCGUCAAAUGGAUGGGAUG__________AGGGGAGG_GGGAUUCGAGCGAUGAGUGCGGCAAC .....(((((((.....)))))))........((((((((((.(((......)))((((((.....))))))......................)))))))..(((.....))))))... (-19.29 = -20.85 + 1.56)

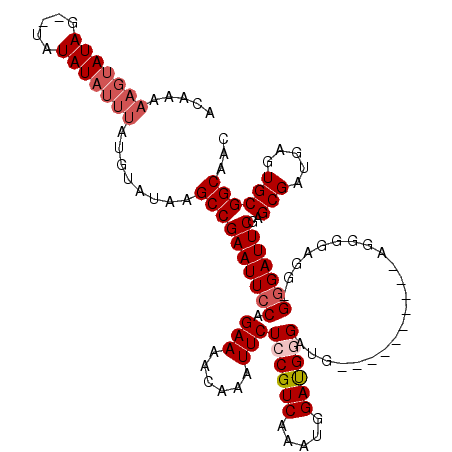

| Location | 14,940,487 – 14,940,606 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.92 |
| Mean single sequence MFE | -28.27 |
| Consensus MFE | -21.39 |
| Energy contribution | -23.20 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.702460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14940487 119 - 27905053 AUGCAACAUUUACGAGCUUGUCGCUGCAAAGAUUUUCAUUCCAGUUCCCCUGCGAGUGGGAGUCUCGGCGAAAAAAUAAGACAA-AAGUAUAGUGUAUAUAUUUAUGUAUAAGCCGAAUU ............((.(((((((((((...(((((..((((((((.....))).)))))..))))))))))).........(((.-(((((((.....))))))).))).))))))).... ( -36.10) >DroSec_CAF1 76301 118 - 1 AUGCAACAUUUACGAGAUUGUCGCUGCAAAGAUUUUCAUUCCAGUUCAGCCGCGAGUGGGAGUCUCGGCGAAAAAAAAAGACAAAAAGUAUAG--UAUAUAUUUAUGUAAAAGCCGAAUU ........((((((......((((((...(((((..((((((.(.....).).)))))..)))))))))))..............(((((((.--..))))))).))))))......... ( -24.90) >DroSim_CAF1 69121 117 - 1 AUGCAACAUUUACGAGCUUGUCGCUGCAAAGAUUUUCAUUCCAGUUCACCCGCGAGUGGGAGUCUCGGCGAAAA-AAAAGACAAAAAGUAUAG--UAUAUAUUUAUGUAUAAGCCGAAUU ............((.(((((((((((...(((((..((((((.........).)))))..)))))))))))...-.....(((..(((((((.--..))))))).))).))))))).... ( -27.70) >DroYak_CAF1 73362 107 - 1 AUGCAACAUUUACGAGCUUGUCGCUGCGAAGAUUUUCAUUCCAGUUCCCC--CGAGUGGGAGUCUCAGGCAGAA-AAAAGACAAAAAGUAUAA--UAUAU--------AUAAGCCGAAUU ............((.(((((((.((((..(((((..(((((.........--.)))))..)))))...))))..-....))).....(((((.--..)))--------)))))))).... ( -24.40) >consensus AUGCAACAUUUACGAGCUUGUCGCUGCAAAGAUUUUCAUUCCAGUUCACCCGCGAGUGGGAGUCUCGGCGAAAA_AAAAGACAAAAAGUAUAG__UAUAUAUUUAUGUAUAAGCCGAAUU ............((.(((((((((((...(((((..(((((............)))))..)))))))))))..............(((((((.....))))))).....))))))).... (-21.39 = -23.20 + 1.81)


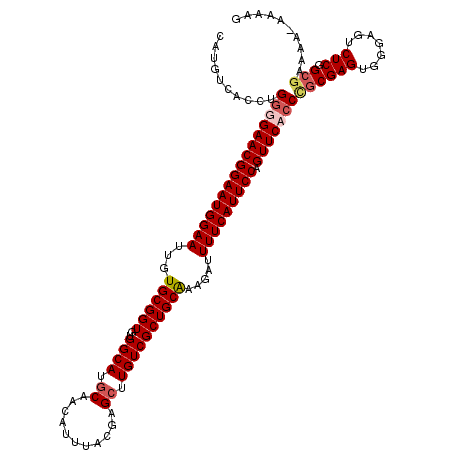
| Location | 14,940,526 – 14,940,646 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -35.20 |
| Consensus MFE | -31.04 |
| Energy contribution | -32.10 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928012 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14940526 120 + 27905053 CUUAUUUUUUCGCCGAGACUCCCACUCGCAGGGGAACUGGAAUGAAAAUCUUUGCAGCGACAAGCUCGUAAAUGUUGCAUGCCACACCGCACAAUUCCAUUCCGUUCCCCAGGUGACAUG .........((((((((.......)))...(((((((.((((((..(((.((((((((.....))).))))).)))...(((......)))......))))))))))))).))))).... ( -39.40) >DroSec_CAF1 76339 120 + 1 CUUUUUUUUUCGCCGAGACUCCCACUCGCGGCUGAACUGGAAUGAAAAUCUUUGCAGCGACAAUCUCGUAAAUGUUGCAUGCCACACCGCACAAUUCCAUUCCGUUCCCCAGGUGACAUG .........((((((((.......)))..((..((((.((((((........(((((((((......))...)))))))(((......)))......)))))))))).)).))))).... ( -30.40) >DroSim_CAF1 69159 119 + 1 CUUUU-UUUUCGCCGAGACUCCCACUCGCGGGUGAACUGGAAUGAAAAUCUUUGCAGCGACAAGCUCGUAAAUGUUGCAUGCCACACCGCACAAUUCCAUUCCGUUCCCCAGGUGACAUG .....-...((((((((.......)))..(((.((((.((((((..(((.((((((((.....))).))))).)))...(((......)))......))))))))))))).))))).... ( -35.70) >DroYak_CAF1 73392 117 + 1 CUUUU-UUCUGCCUGAGACUCCCACUCG--GGGGAACUGGAAUGAAAAUCUUCGCAGCGACAAGCUCGUAAAUGUUGCAUGCCACACCGCACAAUUCCAUUCCGUUCCCCAGGUGACAUG .....-....(((((((.......))).--(((((((.((((((............((((((..........)))))).(((......)))......)))))))))))))))))...... ( -35.30) >consensus CUUUU_UUUUCGCCGAGACUCCCACUCGCGGGGGAACUGGAAUGAAAAUCUUUGCAGCGACAAGCUCGUAAAUGUUGCAUGCCACACCGCACAAUUCCAUUCCGUUCCCCAGGUGACAUG .........((((((((.......)))...(((((((.((((((............((((((..........)))))).(((......)))......))))))))))))).))))).... (-31.04 = -32.10 + 1.06)



| Location | 14,940,526 – 14,940,646 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -39.60 |
| Consensus MFE | -33.79 |
| Energy contribution | -34.98 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.85 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14940526 120 - 27905053 CAUGUCACCUGGGGAACGGAAUGGAAUUGUGCGGUGUGGCAUGCAACAUUUACGAGCUUGUCGCUGCAAAGAUUUUCAUUCCAGUUCCCCUGCGAGUGGGAGUCUCGGCGAAAAAAUAAG ....((.((.((((((((((((((((...((((((..((((.((...........)).)))))))))).....))))))))).)))))))...(((.......))))).))......... ( -42.90) >DroSec_CAF1 76339 120 - 1 CAUGUCACCUGGGGAACGGAAUGGAAUUGUGCGGUGUGGCAUGCAACAUUUACGAGAUUGUCGCUGCAAAGAUUUUCAUUCCAGUUCAGCCGCGAGUGGGAGUCUCGGCGAAAAAAAAAG (((((((((((.(.((..........)).).))).)))))))).................((((((...(((((..((((((.(.....).).)))))..)))))))))))......... ( -33.60) >DroSim_CAF1 69159 119 - 1 CAUGUCACCUGGGGAACGGAAUGGAAUUGUGCGGUGUGGCAUGCAACAUUUACGAGCUUGUCGCUGCAAAGAUUUUCAUUCCAGUUCACCCGCGAGUGGGAGUCUCGGCGAAAA-AAAAG ....((.((.((((((((((((((((...((((((..((((.((...........)).)))))))))).....))))))))).)))).)))..(((.......))))).))...-..... ( -39.60) >DroYak_CAF1 73392 117 - 1 CAUGUCACCUGGGGAACGGAAUGGAAUUGUGCGGUGUGGCAUGCAACAUUUACGAGCUUGUCGCUGCGAAGAUUUUCAUUCCAGUUCCCC--CGAGUGGGAGUCUCAGGCAGAA-AAAAG ..((((....((((((((((((((((...((((((..((((.((...........)).)))))))))).....))))))))).)))))))--.(((.......))).))))...-..... ( -42.30) >consensus CAUGUCACCUGGGGAACGGAAUGGAAUUGUGCGGUGUGGCAUGCAACAUUUACGAGCUUGUCGCUGCAAAGAUUUUCAUUCCAGUUCACCCGCGAGUGGGAGUCUCGGCGAAAA_AAAAG ..........((((((((((((((((...((((((..((((.((...........)).)))))))))).....))))))))).)))))))((((((.......))).))).......... (-33.79 = -34.98 + 1.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:52 2006