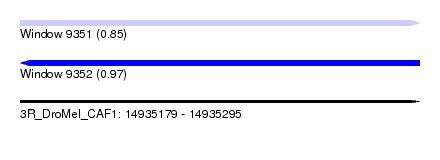
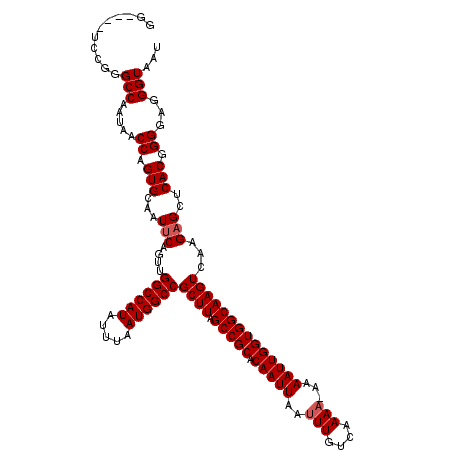
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,935,179 – 14,935,295 |
| Length | 116 |
| Max. P | 0.965846 |

| Location | 14,935,179 – 14,935,295 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.47 |
| Mean single sequence MFE | -30.32 |
| Consensus MFE | -26.32 |
| Energy contribution | -26.72 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14935179 116 + 27905053 AUUACCCUCCCCGUGAGCUCUUGACUUGCCACCAAUUUUUUUUUGACAAAUUAAUUGUGCGGCUAAGCGGCCAUUAAAUAUGGCCAACUGUAUUGCACUGGUUAUUGGCCCGGA----CC ..........(((.(.(((..(((((.(((.((((((..(((.....)))..))))).).))).....((((((.....))))))..............)))))..))))))).----.. ( -30.70) >DroSec_CAF1 66628 115 + 1 AUUACCCUCCCCGUGAGCUCUUGACUUGCCACCAAUUUU-UUUUGACAAAUUAAUUGUGCGGCUAAGCGGCCAUUAAAUAUGGCCAACUGAAUUGCACUGGUUAUUGGCCCGGA----CC ..........(((.(.(((..(((((.(((.((((((..-(((....)))..))))).).))).....((((((.....))))))..............)))))..))))))).----.. ( -30.70) >DroSim_CAF1 63705 115 + 1 AUUACCCUCCCCGUGAGCUCUUGACUUGCCACCAAUUUU-UUUUGACAAAUUAAUUGUGCGGCUAAGCGGCCAUUAAAUAUGGCCAACUGAAUUGCACUGGUUAUUGGCCCGGA----CC ..........(((.(.(((..(((((.(((.((((((..-(((....)))..))))).).))).....((((((.....))))))..............)))))..))))))).----.. ( -30.70) >DroEre_CAF1 61591 117 + 1 AUUACCCUCCCCGUGAGCUCUUGACUUGCCACCAAUUU---UUUGACAAAUUAAUUGUGCGGCUAAGCGGCCAUUAAAUAUGGCCAACUGAAUUGCACUGGUUAUUGGCCCGGGCCGUCC .........((((.(.(((..(((((.(((.((((((.---.((....))..))))).).))).....((((((.....))))))..............)))))..))))))))...... ( -33.70) >DroYak_CAF1 68439 114 + 1 ACUACCCUCCCCGUGAACUCUUGACUUGCCACCAAUUU--UUUUGACAAAUUAAUUGUGCGGCUAAGCGGCCAUUAAAUAUGGCCAACUGAAUUGCACUGGUUAUUGGCCCGUC----CC ......................(((..((((.(((...--..)))......(((((((((((...((.((((((.....))))))..))...)))))).))))).))))..)))----.. ( -25.80) >consensus AUUACCCUCCCCGUGAGCUCUUGACUUGCCACCAAUUUU_UUUUGACAAAUUAAUUGUGCGGCUAAGCGGCCAUUAAAUAUGGCCAACUGAAUUGCACUGGUUAUUGGCCCGGA____CC ..........(((.(.(((..(((((.(((.((((((...............))))).).))).....((((((.....))))))..............)))))..)))))))....... (-26.32 = -26.72 + 0.40)

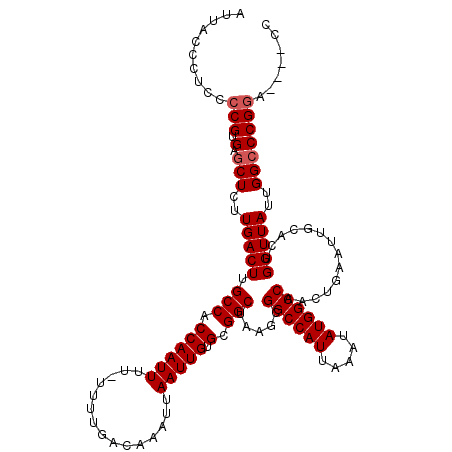

| Location | 14,935,179 – 14,935,295 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.47 |
| Mean single sequence MFE | -34.18 |
| Consensus MFE | -31.50 |
| Energy contribution | -31.90 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14935179 116 - 27905053 GG----UCCGGGCCAAUAACCAGUGCAAUACAGUUGGCCAUAUUUAAUGGCCGCUUAGCCGCACAAUUAAUUUGUCAAAAAAAAAUUGGUGGCAAGUCAAGAGCUCACGGGGAGGGUAAU ((----(....)))....(((..(.(.....((((((((((.....))))))((((.(((((.(((((..(((.....)))..))))))))))))))....))))....).)..)))... ( -32.90) >DroSec_CAF1 66628 115 - 1 GG----UCCGGGCCAAUAACCAGUGCAAUUCAGUUGGCCAUAUUUAAUGGCCGCUUAGCCGCACAAUUAAUUUGUCAAAA-AAAAUUGGUGGCAAGUCAAGAGCUCACGGGGAGGGUAAU ((----(....))).....((.(((...(((....((((((.....))))))((((.(((((.(((((..(((....)))-..))))))))))))))...)))..))).))......... ( -33.30) >DroSim_CAF1 63705 115 - 1 GG----UCCGGGCCAAUAACCAGUGCAAUUCAGUUGGCCAUAUUUAAUGGCCGCUUAGCCGCACAAUUAAUUUGUCAAAA-AAAAUUGGUGGCAAGUCAAGAGCUCACGGGGAGGGUAAU ((----(....))).....((.(((...(((....((((((.....))))))((((.(((((.(((((..(((....)))-..))))))))))))))...)))..))).))......... ( -33.30) >DroEre_CAF1 61591 117 - 1 GGACGGCCCGGGCCAAUAACCAGUGCAAUUCAGUUGGCCAUAUUUAAUGGCCGCUUAGCCGCACAAUUAAUUUGUCAAA---AAAUUGGUGGCAAGUCAAGAGCUCACGGGGAGGGUAAU .....((((((........)).(((...(((....((((((.....))))))((((.(((((.(((((...........---.))))))))))))))...)))..))).....))))... ( -35.00) >DroYak_CAF1 68439 114 - 1 GG----GACGGGCCAAUAACCAGUGCAAUUCAGUUGGCCAUAUUUAAUGGCCGCUUAGCCGCACAAUUAAUUUGUCAAAA--AAAUUGGUGGCAAGUCAAGAGUUCACGGGGAGGGUAGU ..----.((..(((.....((.(((.(((((....((((((.....))))))((((.(((((.(((((..(((.....))--)))))))))))))))...)))))))).))...))).)) ( -36.40) >consensus GG____UCCGGGCCAAUAACCAGUGCAAUUCAGUUGGCCAUAUUUAAUGGCCGCUUAGCCGCACAAUUAAUUUGUCAAAA_AAAAUUGGUGGCAAGUCAAGAGCUCACGGGGAGGGUAAU ...........(((.....((.(((...(((....((((((.....))))))((((.(((((.(((((..(((....)))...))))))))))))))...)))..))).))...)))... (-31.50 = -31.90 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:39 2006