
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,934,986 – 14,935,146 |
| Length | 160 |
| Max. P | 0.919951 |

| Location | 14,934,986 – 14,935,106 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -30.57 |
| Consensus MFE | -26.58 |
| Energy contribution | -26.66 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.87 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14934986 120 + 27905053 UCUACGGCUAAAUAAAUGCGAGGCAUCUGCAAAGUGUCUAAUUAAAUGUGCCGAAUUUAAUGAUGAUAACUCUGGCUAAGACCAUUGCCUGCAGUGCAAUUUUGGCCGGCUCUGACUUUA ....((((((((....((((..(((...((((..((((..(((((((.......)))))))...))))....(((......))))))).)))..))))..))))))))............ ( -30.80) >DroSec_CAF1 66435 120 + 1 UCUACGGCUAAAUAAAUGCGAGGCAUCUGCAAAGUGUCUAAUUAAAUGUGCCGAAUUUAAUGAUGAUAACUCUGGCUUAGACCAUUGCCUGCAGUGCAAUCUUGGCCGGCUCUGACUUUA ....(((((((.....((((..(((...((((.(.((((((........((((...................))))))))))).)))).)))..))))...)))))))............ ( -29.31) >DroSim_CAF1 63520 120 + 1 UCUACGGCCAAAUAAAUGCGAGGCAUCUGCAAAGUGUCUAAUUAAAUGUGCCGAAUUUAAUGAUGAUAACUCUGGCUUAGACCAUUGCCUGCAGUGCAAUCUUGGCCGGCUCUGACUCUA ....(((((((.....((((..(((...((((.(.((((((........((((...................))))))))))).)))).)))..))))...)))))))............ ( -31.61) >DroEre_CAF1 61399 119 + 1 UCUCCGGCUAAAUAAAUGCGAGGCAUCUGCAAAGUGUCUAAUUAAAUGUGCCGAAUUUAAUGAUGAUAACUUUGGCUUAGA-CAUUGCCUGCAGUUCAAUCUUGGCCGGCUCUGACUUUU ...((((((((.....(((.((((((((((((((((((..(((((((.......)))))))...))).))))))...))))-...))))))))........))))))))........... ( -34.32) >DroYak_CAF1 68240 119 + 1 UCUCCAGCUAAAUAAAUGCGAGGUAUCUGCAAAGUGUCUAAUUAAAUGUGCCGAAUUUAAUGAUGAUAACUUUGGCUUAGA-CAUUGCCUGCAGUUCAAUUUUGCCCGGCUCUGACUUUU ((...((((........(((((((..(((((.(((((((((........((((((.(((.......))).)))))))))))-))))...)))))....)))))))..))))..))..... ( -26.80) >consensus UCUACGGCUAAAUAAAUGCGAGGCAUCUGCAAAGUGUCUAAUUAAAUGUGCCGAAUUUAAUGAUGAUAACUCUGGCUUAGACCAUUGCCUGCAGUGCAAUCUUGGCCGGCUCUGACUUUA ....(((((((.....(((.((((((((((..((((((..(((((((.......)))))))...))).)))...))..)))....))))))))........)))))))............ (-26.58 = -26.66 + 0.08)


| Location | 14,935,026 – 14,935,146 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.16 |
| Mean single sequence MFE | -34.54 |
| Consensus MFE | -32.26 |
| Energy contribution | -32.82 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14935026 120 + 27905053 AUUAAAUGUGCCGAAUUUAAUGAUGAUAACUCUGGCUAAGACCAUUGCCUGCAGUGCAAUUUUGGCCGGCUCUGACUUUAGCAUUUUGGCCAACCGGUCAAGGUGAAAGUUGCGCCACCU (((((((.......)))))))............((((((((.(((((....)))))...))))))))(((.(.(((((...((((((((((....)))))))))).)))))).))).... ( -37.10) >DroSec_CAF1 66475 120 + 1 AUUAAAUGUGCCGAAUUUAAUGAUGAUAACUCUGGCUUAGACCAUUGCCUGCAGUGCAAUCUUGGCCGGCUCUGACUUUAGCAUUUUGGCCAACCGGUCAAGGUGAAAGUUGCGCCACCU (((((((.......)))))))............((((.(((.(((((....)))))...))).))))(((.(.(((((...((((((((((....)))))))))).)))))).))).... ( -36.10) >DroSim_CAF1 63560 119 + 1 AUUAAAUGUGCCGAAUUUAAUGAUGAUAACUCUGGCUUAGACCAUUGCCUGCAGUGCAAUCUUGGCCGGCUCUGACUCUAGCAUUUUGGCCAACCGGUCAAGGUGAAAGUUGCGCCACC- (((((((.......)))))))............((((.(((.(((((....)))))...))).))))(((.(.((((....((((((((((....))))))))))..))))).)))...- ( -34.90) >DroEre_CAF1 61439 119 + 1 AUUAAAUGUGCCGAAUUUAAUGAUGAUAACUUUGGCUUAGA-CAUUGCCUGCAGUUCAAUCUUGGCCGGCUCUGACUUUUGCAUUUUGGCCAACCGGUCAAGGUGAAAGUUGCGCCACCU .......(.(((((.......(((...((((..(((.....-....)))...))))..)))))))))(((.(.((((((..(...((((((....)))))).)..))))))).))).... ( -33.11) >DroYak_CAF1 68280 119 + 1 AUUAAAUGUGCCGAAUUUAAUGAUGAUAACUUUGGCUUAGA-CAUUGCCUGCAGUUCAAUUUUGCCCGGCUCUGACUUUUGCAUUUUGGCCAACCGGUCAAGGUGAAAGUUGCGCCACCU ....(((((((((((.(((.......))).))))))....)-))))....((((.......))))..(((.(.((((((..(...((((((....)))))).)..))))))).))).... ( -31.50) >consensus AUUAAAUGUGCCGAAUUUAAUGAUGAUAACUCUGGCUUAGACCAUUGCCUGCAGUGCAAUCUUGGCCGGCUCUGACUUUAGCAUUUUGGCCAACCGGUCAAGGUGAAAGUUGCGCCACCU (((((((.......)))))))............((((.(((.(((((....)))))...))).))))(((.(.(((((...((((((((((....)))))))))).)))))).))).... (-32.26 = -32.82 + 0.56)

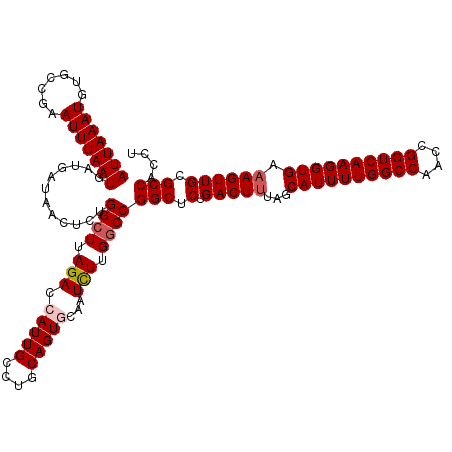
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:37 2006