
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,934,702 – 14,934,897 |
| Length | 195 |
| Max. P | 0.786084 |

| Location | 14,934,702 – 14,934,807 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.11 |
| Mean single sequence MFE | -30.58 |
| Consensus MFE | -20.30 |
| Energy contribution | -19.98 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.731654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14934702 105 - 27905053 UUUUUCGCAUUUUUGUGCGCUGUUAAUUAGCCACGAGAAGCGUAAUGCGCAUAAUGGCUAUUAGAUGGUAUAGCAGAUGUA-------UAUAUAGAUAGAU------AGAU--AUCGGAC .....(((.((((((((.((((.....)))))))))))))))..(((..(((....(((((........)))))..)))..-------)))...((((...------...)--))).... ( -27.20) >DroSec_CAF1 64580 107 - 1 UUUUUCGCAUUUUUGUGCGCUGUUAAUUAGCCACGAGAAGCGUAAUGCGCAUAAUGGCCAUUAGAUGGUAUGGCAGAUGUA-------UAUAUGGAUAGAU------AGAUAUAUCGGAC .....(((.((((((((.((((.....)))))))))))))))...(((.(((....(((((...)))))))))))((((((-------(.(((......))------).))))))).... ( -29.30) >DroSim_CAF1 63250 107 - 1 UUUUUCGCAUUUUUGUGCGCUGUUAAUUAGCCACGAGAAGCGUAAUGUGCAUAAUGGCCAUUAGAUGGUAUAGCAGAUGUA-------UGUAUGGAUGUAU------AGAUAUAUCGGAC .....(((.((((((((.((((.....))))))))))))))).....(((.((...(((((...))))).)))))((((((-------(.((((....)))------).))))))).... ( -30.20) >DroEre_CAF1 61116 117 - 1 UUUUUCGCAUUUGUGUGCGCUGUUAAUUAGCCGCGAGAAGCGUAAUGCGCAUAAUGGCUAUUAGAUGGUAUGGAAGAU--AAGC-UCGUAUAUGAAAAGAUAUGCCCAGGCAUAUCGGAG .(((((((..((((((((((((.....))))(((.....)))....))))))))..)).........((((((.....--....-))))))..)))))(((((((....))))))).... ( -34.30) >DroYak_CAF1 67953 120 - 1 UUUUUCGCAUUUUUGUGCGCUGUUAAUUAGCCACGAGAAGCGUAAUGAGCAUAAUGGCUAUUAGAUGGUGUGGAAGAAAUAUGUAUGUUAUAUGGAGAGAUAUGCUCAGGUAUAUCGGAG .(((((((.((((((((.((((.....))))))))))))))((((((.(((((....((((........))))......))))).))))))..)))))(((((((....))))))).... ( -31.90) >consensus UUUUUCGCAUUUUUGUGCGCUGUUAAUUAGCCACGAGAAGCGUAAUGCGCAUAAUGGCUAUUAGAUGGUAUGGCAGAUGUA_______UAUAUGGAUAGAU______AGAUAUAUCGGAC .....(((.((((((((.((((.....)))))))))))))))(((((.((......)))))))..(((((((......................................)))))))... (-20.30 = -19.98 + -0.32)



| Location | 14,934,727 – 14,934,847 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.74 |
| Mean single sequence MFE | -30.06 |
| Consensus MFE | -25.66 |
| Energy contribution | -25.90 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.786084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14934727 120 - 27905053 AAUUGGACAAAAAGACAACUGCGGAGAAGGCAUUGAAAAGUUUUUCGCAUUUUUGUGCGCUGUUAAUUAGCCACGAGAAGCGUAAUGCGCAUAAUGGCUAUUAGAUGGUAUAGCAGAUGU ..............(((.((((...(((((.(((....))))))))((((....))))((((((...((((((..(...(((.....))).)..))))))...))))))...)))).))) ( -30.40) >DroSec_CAF1 64607 120 - 1 AAUUGGACAAAAAGACAACUGCGAAGAAGGCAUUGAAAAGUUUUUCGCAUUUUUGUGCGCUGUUAAUUAGCCACGAGAAGCGUAAUGCGCAUAAUGGCCAUUAGAUGGUAUGGCAGAUGU ..............(((.((((.......((((((....(((((((((((....))))((((.....))))...))))))).))))))........(((((...)))))...)))).))) ( -32.50) >DroSim_CAF1 63277 120 - 1 AAUUGGACAAAAAGACAACUGCGCAGAAGGCAUUGAAAAGUUUUUCGCAUUUUUGUGCGCUGUUAAUUAGCCACGAGAAGCGUAAUGUGCAUAAUGGCCAUUAGAUGGUAUAGCAGAUGU ..............(((.(((((((.(..((...((((....))))((.((((((((.((((.....))))))))))))))))..).)))......(((((...)))))...)))).))) ( -33.10) >DroEre_CAF1 61155 116 - 1 AAUUGGACAG--AGACAACUGCAAAGAAGGCAUUGAAAAGUUUUUCGCAUUUGUGUGCGCUGUUAAUUAGCCGCGAGAAGCGUAAUGCGCAUAAUGGCUAUUAGAUGGUAUGGAAGAU-- ..........--...(((.(((.......))))))....(((((((((((....))))((((((...((((((..(...(((.....))).)..))))))...))))))..)))))))-- ( -25.80) >DroYak_CAF1 67993 118 - 1 AAUUGGACAG--AGACAUCUGCAAAGAAGGCAUUGAAAAGUUUUUCGCAUUUUUGUGCGCUGUUAAUUAGCCACGAGAAGCGUAAUGAGCAUAAUGGCUAUUAGAUGGUGUGGAAGAAAU .......((.--(..((((((......((.(((((....((((..(((.((((((((.((((.....)))))))))))))))....)))).))))).))..)))))).).))........ ( -28.50) >consensus AAUUGGACAAAAAGACAACUGCGAAGAAGGCAUUGAAAAGUUUUUCGCAUUUUUGUGCGCUGUUAAUUAGCCACGAGAAGCGUAAUGCGCAUAAUGGCUAUUAGAUGGUAUGGCAGAUGU ..................((((.......((((((....(((((((((((....))))((((.....))))...))))))).))))))........(((((...)))))...)))).... (-25.66 = -25.90 + 0.24)

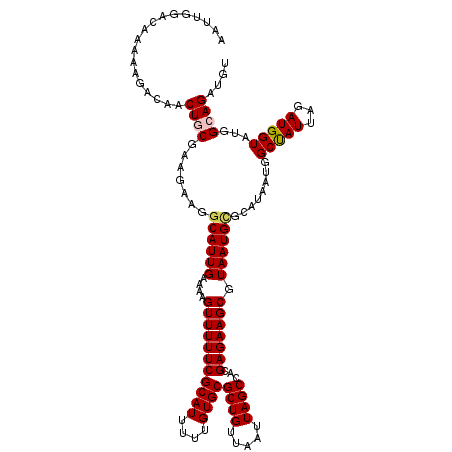

| Location | 14,934,807 – 14,934,897 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.39 |
| Mean single sequence MFE | -15.41 |
| Consensus MFE | -11.45 |
| Energy contribution | -12.74 |
| Covariance contribution | 1.29 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.628600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14934807 90 + 27905053 CUUUUCAAUGCCUUCUCCGCAGUUGUCUUUUUGUCCAAUUUGCCAAGCUUAGAAAUGCAAAUUACCAAAAGCAGCGAAAAUAAAAAAAAC------------------------------ .(((((..(((.......)))((((.(((((.((..(((((((.............))))))))).))))))))))))))..........------------------------------ ( -15.92) >DroSec_CAF1 64687 120 + 1 CUUUUCAAUGCCUUCUUCGCAGUUGUCUUUUUGUCCAAUUUGCCAAGCUUAGAAAUGCAAAUUACCAAAAGCAGCGAAAAUAAAAAACACAAACUCAAACAAAACGCGAACAAAGAGCUA .........(((((.(((((.((((.(((((.((..(((((((.............))))))))).)))))))))((.................)).........)))))..))).)).. ( -19.85) >DroSim_CAF1 63357 92 + 1 CUUUUCAAUGCCUUCUGCGCAGUUGUCUUUUUGUCCAAUUUGCCAAGCUUAGAAAUGCAAAUUACCAAAAGCAGCGAAAAUAAAAAACACAA---------------------------- .(((((..((((....).)))((((.(((((.((..(((((((.............))))))))).))))))))))))))............---------------------------- ( -16.82) >DroEre_CAF1 61233 81 + 1 CUUUUCAAUGCCUUCUUUGCAGUUGUCU--CUGUCCAAUUUGCCAAGCUUAGAAAUGCAAAUGACCAA-AGCAGCGAAAAUAA--C---------------------------------- .(((((..(((.......)))((((..(--..(((..((((((.............)))))))))..)-..)))))))))...--.---------------------------------- ( -14.12) >DroYak_CAF1 68073 82 + 1 CUUUUCAAUGCCUUCUUUGCAGAUGUCU--CUGUCCAAUUUGCCAAGCUUAGAAAUGCAAAUUACCAAAAACAGCGAAAAAAA--U---------------------------------- .(((((............(((((....)--))))..(((((((.............)))))))............)))))...--.---------------------------------- ( -10.32) >consensus CUUUUCAAUGCCUUCUUCGCAGUUGUCUUUUUGUCCAAUUUGCCAAGCUUAGAAAUGCAAAUUACCAAAAGCAGCGAAAAUAAAAAA_AC______________________________ .(((((..(((.......)))(((((..(((((...(((((((.............)))))))..)))))))))))))))........................................ (-11.45 = -12.74 + 1.29)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:35 2006