
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,934,080 – 14,934,195 |
| Length | 115 |
| Max. P | 0.997523 |

| Location | 14,934,080 – 14,934,195 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.75 |
| Mean single sequence MFE | -27.62 |
| Consensus MFE | -23.67 |
| Energy contribution | -24.11 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.635696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
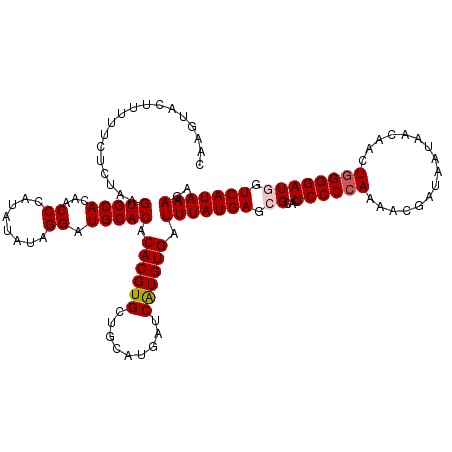
>3R_DroMel_CAF1 14934080 115 + 27905053 -----ACUUGUUCUCUAAGUGCACAACCCAUAUUUAGGAUGCACACACGUGCUGCAUGAUCAUGUGAUUUAUGAGCGCACUCCCCAAAACGAUAAUAACAACUAGGGAUGGUCAUAAAGA -----(((...((((((.(((((...((........)).)))))....(((((.(((((((....)).))))))))))........................)))))).)))........ ( -24.80) >DroSec_CAF1 63974 120 + 1 CAAGUACUUUUUCUCUAAGUGCACAACCCAUAUCUAGGAUGCACACACGUGCUGCAUGAUCAUGUGAUUUAUGAGCGCACUCCCCAAAACGAUAAUAACAACUGGGGAUGGUCAUAAAGA ......((((....(((.(((((...((........)).)))))....(((((.(((((((....)).))))))))))..((((((................)))))))))....)))). ( -29.09) >DroSim_CAF1 62644 120 + 1 CAAGUAUUUUUUCUCUAAGUGCACAACCCAUAUCUAGGAUGCACACACGUGCUGCAUGAUCAUGUGAUUUAUGAGCGCACUCCCCAAAACGAUAAUGACAACUGGGGAUGGUCAUAAAGA ..................(((((...((........)).))))).((((((.........)))))).(((((((.((...((((((................)))))))).))))))).. ( -27.29) >DroEre_CAF1 60500 120 + 1 CAAAUAUAUUUUCUCUAAGUGCACAACCCAUAUAUAGGAUGCACACACGUGCUGCAUGAUCAUGUGAUUUAUGACCGCACUCCCCAAAACGGUAAUAACAACUGGGGAUUGUCAUAAAGA ..................(((((...((........)).))))).((((((.........)))))).((((((((.....((((((.....((....))...))))))..)))))))).. ( -27.60) >DroYak_CAF1 67350 120 + 1 CAACUACAUUUUCUCUAAGUGCACAGCCCAUAUAUAGGAUGCACACACGUGCUGCAUGAUCGUGUGAUUUAUGACCGCACUCCCCAAAACGGUAAUAACUACUGGGGAUUGUCAUAAAGA ..................(((((...((........)).)))))(((((..(.....)..)))))..((((((((.....((((((....(((....)))..))))))..)))))))).. ( -29.30) >consensus CAAGUACUUUUUCUCUAAGUGCACAACCCAUAUAUAGGAUGCACACACGUGCUGCAUGAUCAUGUGAUUUAUGAGCGCACUCCCCAAAACGAUAAUAACAACUGGGGAUGGUCAUAAAGA ..................(((((...((........)).))))).((((((.........)))))).(((((((.((...((((((................)))))))).))))))).. (-23.67 = -24.11 + 0.44)


| Location | 14,934,080 – 14,934,195 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.75 |
| Mean single sequence MFE | -34.06 |
| Consensus MFE | -33.24 |
| Energy contribution | -33.08 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.88 |
| SVM RNA-class probability | 0.997523 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
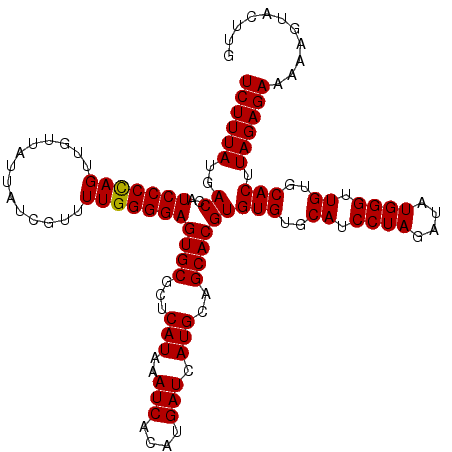
>3R_DroMel_CAF1 14934080 115 - 27905053 UCUUUAUGACCAUCCCUAGUUGUUAUUAUCGUUUUGGGGAGUGCGCUCAUAAAUCACAUGAUCAUGCAGCACGUGUGUGCAUCCUAAAUAUGGGUUGUGCACUUAGAGAACAAGU----- ((((((......(((((((.((.......))..)))))))((((((((((.......))))....)).))))..(((..((.((((....)))).))..))).))))))......----- ( -31.00) >DroSec_CAF1 63974 120 - 1 UCUUUAUGACCAUCCCCAGUUGUUAUUAUCGUUUUGGGGAGUGCGCUCAUAAAUCACAUGAUCAUGCAGCACGUGUGUGCAUCCUAGAUAUGGGUUGUGCACUUAGAGAAAAAGUACUUG ((((((......(((((((.((.......))..)))))))((((((((((.......))))....)).))))..(((..((.((((....)))).))..))).))))))........... ( -34.10) >DroSim_CAF1 62644 120 - 1 UCUUUAUGACCAUCCCCAGUUGUCAUUAUCGUUUUGGGGAGUGCGCUCAUAAAUCACAUGAUCAUGCAGCACGUGUGUGCAUCCUAGAUAUGGGUUGUGCACUUAGAGAAAAAAUACUUG ((((((......(((((((.((.......))..)))))))((((((((((.......))))....)).))))..(((..((.((((....)))).))..))).))))))........... ( -34.10) >DroEre_CAF1 60500 120 - 1 UCUUUAUGACAAUCCCCAGUUGUUAUUACCGUUUUGGGGAGUGCGGUCAUAAAUCACAUGAUCAUGCAGCACGUGUGUGCAUCCUAUAUAUGGGUUGUGCACUUAGAGAAAAUAUAUUUG ((((((......(((((((..((....))....)))))))((((((((((.......))))))..(((((.((((((((.....)))))))).))))))))).))))))........... ( -35.70) >DroYak_CAF1 67350 120 - 1 UCUUUAUGACAAUCCCCAGUAGUUAUUACCGUUUUGGGGAGUGCGGUCAUAAAUCACACGAUCAUGCAGCACGUGUGUGCAUCCUAUAUAUGGGCUGUGCACUUAGAGAAAAUGUAGUUG ((((((......(((((((..((....))....)))))))((((((((...........))))..(((((.((((((((.....)))))))).))))))))).))))))........... ( -35.40) >consensus UCUUUAUGACCAUCCCCAGUUGUUAUUAUCGUUUUGGGGAGUGCGCUCAUAAAUCACAUGAUCAUGCAGCACGUGUGUGCAUCCUAGAUAUGGGUUGUGCACUUAGAGAAAAAGUACUUG ((((((..((..(((((((..............)))))))((((...(((..(((....))).)))..))))))(((..((.((((....)))).))..))).))))))........... (-33.24 = -33.08 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:31 2006