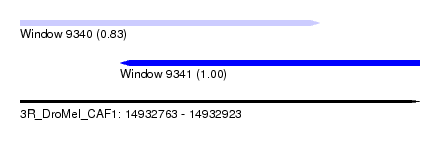
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,932,763 – 14,932,923 |
| Length | 160 |
| Max. P | 0.999171 |

| Location | 14,932,763 – 14,932,883 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.00 |
| Mean single sequence MFE | -31.92 |
| Consensus MFE | -28.80 |
| Energy contribution | -29.04 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14932763 120 + 27905053 UACAACGAAGGCUAAGGCCAUCCGCGGAGAAAGAGACGGCAUGCGUGGGCGCUGUUAAGUCAAUGUUAAAAGCGCAUGUUGGGCCAAUGUUAAGUGCUAAUAAGUAAACACUGCAAAAAA ......((.(((....))).)).((((.........((((((((((.((((.((......)).))))....))))))))))......((((...((((....))))))))))))...... ( -31.20) >DroSec_CAF1 62633 120 + 1 UACAACGAAGGCUAAGGCCAUCCGCGGAGAAAGAGACGGCAUGCGUGGGCGCUGUUAAGUCAAUGUUAAAAGCGCAUGUUGGGCCAAUGUUAAGUGCUAAUAAGUAAACACUGCAAAAAA ......((.(((....))).)).((((.........((((((((((.((((.((......)).))))....))))))))))......((((...((((....))))))))))))...... ( -31.20) >DroSim_CAF1 61332 120 + 1 UACAACGAAGGCUAAGGCCAUCCGCGGAGAAAGAGACGGCAUGCGUGGGCGCUGUUAAGUCAAUGUUAAAAGCGCAUGUUGGGCCAAUGUUAAGUGCUAAUAAGUAAACACUGCAAAAAA ......((.(((....))).)).((((.........((((((((((.((((.((......)).))))....))))))))))......((((...((((....))))))))))))...... ( -31.20) >DroEre_CAF1 59182 119 + 1 UACAACGAAGGCUAAGGCCAUCCGCGGAGAAAGAGACGGCAUGCGUGGGCGCUGUUAAGUCAAUGUUA-AAGCGCAUGUUGGGCCAAUGUUAAGUGCUAAUAAGUAAACACUGCAAAAAA ......((.(((....))).)).((((.........((((((((((.((((.((......)).)))).-..))))))))))......((((...((((....))))))))))))...... ( -31.80) >DroYak_CAF1 66176 119 + 1 UACAACGAAGGCUAAGGCCAUCCGCGGAGAAAGAGACGGCGUGCGUGGGCGCUGUUAAGUCAAUGUUAAAAGCGCAGGCUG-GCCAAUACUUAGUGCUAAUAAGUAAACACUCCAAAAAA ......((.(((....))).))...((((.......((((.(((((.((((.((......)).))))....))))).))))-.....((((((.......))))))....))))...... ( -34.20) >consensus UACAACGAAGGCUAAGGCCAUCCGCGGAGAAAGAGACGGCAUGCGUGGGCGCUGUUAAGUCAAUGUUAAAAGCGCAUGUUGGGCCAAUGUUAAGUGCUAAUAAGUAAACACUGCAAAAAA ......((.(((....))).)).((((.........((((((((((.((((.((......)).))))....)))))))))).............((((....))))....))))...... (-28.80 = -29.04 + 0.24)



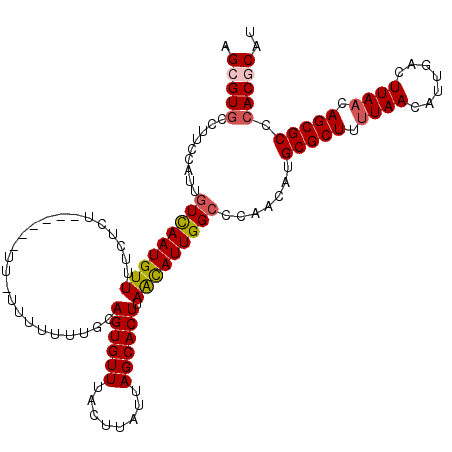
| Location | 14,932,803 – 14,932,923 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.11 |
| Mean single sequence MFE | -25.06 |
| Consensus MFE | -21.83 |
| Energy contribution | -22.35 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.41 |
| SVM RNA-class probability | 0.999171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14932803 120 - 27905053 AGCGUGCCUUCCAUUGUCAAUGUUUUCUCUAUUCUCUUUUUUUUUUGCAGUGUUUACUUAUUAGCACUUAACAUUGGCCCAACAUGCGCUUUUAACAUUGACUUAACAGCGCCCACGCAU .(((((.........(((((((((........................((((((........)))))).))))))))).......(((((.((((.......)))).))))).))))).. ( -27.51) >DroSec_CAF1 62673 115 - 1 AGCGUGUCUUCCAUUGUCAAUGUUUUCUCUA-----UUUUUUUUUUGCAGUGUUUACUUAUUAGCACUUAACAUUGGCCCAACAUGCGCUUUUAACAUUGACUUAACAGCGCCCACGCAU .(((((.........(((((((((.......-----............((((((........)))))).))))))))).......(((((.((((.......)))).))))).))))).. ( -27.95) >DroSim_CAF1 61372 114 - 1 AGCGUGCCUU-CAUUGUCAAUGUUUUCUCUC-----UUCUUUUUUUGCAGUGUUUACUUAUUAGCACUUAACAUUGGCCCAACAUGCGCUUUUAACAUUGACUUAACAGCGCCCACGCAU .(((((....-....(((((((((.......-----............((((((........)))))).))))))))).......(((((.((((.......)))).))))).))))).. ( -27.75) >DroEre_CAF1 59222 110 - 1 AGAGUGUCUUCCAUUGUCAAUGUUUUCUCU---------UUUUUUUGCAGUGUUUACUUAUUAGCACUUAACAUUGGCCCAACAUGCGCUU-UAACAUUGACUUAACAGCGCCCACGCAU ...((((........(((((((((......---------.........((((((........)))))).))))))))).......((((((-(((.......)))).)))))..)))).. ( -21.99) >DroYak_CAF1 66216 108 - 1 AGAGUUCAUUUCAUU-UUAAUGUUUUCUU----------UUUUUUUGGAGUGUUUACUUAUUAGCACUAAGUAUUGGC-CAGCCUGCGCUUUUAACAUUGACUUAACAGCGCCCACGCAC ((((..((((.....-..))))..)))).----------......(((((((((........)))))).......((.-...)).(((((.((((.......)))).))))))))..... ( -20.10) >consensus AGCGUGCCUUCCAUUGUCAAUGUUUUCUCU______UU_UUUUUUUGCAGUGUUUACUUAUUAGCACUUAACAUUGGCCCAACAUGCGCUUUUAACAUUGACUUAACAGCGCCCACGCAU .(((((.........(((((((((........................((((((........)))))).))))))))).......(((((.((((.......)))).))))).))))).. (-21.83 = -22.35 + 0.52)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:28 2006