
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,932,183 – 14,932,336 |
| Length | 153 |
| Max. P | 0.746330 |

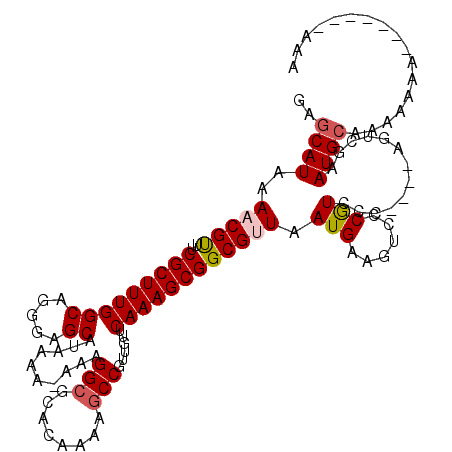
| Location | 14,932,183 – 14,932,299 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.14 |
| Mean single sequence MFE | -25.48 |
| Consensus MFE | -19.82 |
| Energy contribution | -20.30 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14932183 116 - 27905053 GAGCAUAAAACGUUUCGCUUUGGCAGGGAGCAUAAAAAAAAGGCGCACAAAAGCCGUUGUUCAAAGCGGCGUUAAUGAAGUCCCGUCCC----AGUCGAAUGCAAAAAAAAAAAAAAAAA ..((((........(((..((((..((((((...........))........((((((......))))))..........))))...))----)).)))))))................. ( -25.40) >DroSec_CAF1 62102 105 - 1 CAGCAUAAAACGUUUCGCUUUGGCAGGGAGCAUAAAA-AAAGGCGCACAAAAGCCGUUGUUCAAAGCGGCGUUAAUGAAGUCCCGUCCC----AGUCGAAUGAAAAAA----------AA ............(((((.((((((.((((((......-....))........((((((......))))))...............))))----.)))))))))))...----------.. ( -25.40) >DroSim_CAF1 60797 106 - 1 CAGCAUAAAACGUUUCGCUUUGGCAGGGAGCAUAAAA-AAAGGCGCACAAAAGCCGUUGUUCAAAGCGGCGUUAAUGAAGUCCCGUCCC----AGUCGAAUGCAAAAA---------AAA ..((((........(((..((((..((((((......-....))........((((((......))))))..........))))...))----)).))))))).....---------... ( -25.50) >DroEre_CAF1 58655 104 - 1 GAGCAUAAAACGUUUCGCUUUGGCAGGGAGCAUAAA--AAAGGCGCACAAAAGCCGUUGUUCAAAGCGGCGAUAAUGAAGUCCCGUCCC----AGUCAAAUGCAAAAAAA---------- ................((((((((.(((((((....--...(((........)))..)))))...((((.(((......))))))).))----.)))))).)).......---------- ( -26.10) >DroYak_CAF1 65653 113 - 1 GAGCAUAAAACGCUUCGCUUUGGCGGAGAGCAUAAA--AAAGGCACACAAAAUCCGUUGUUCAAAGCGGCGAUAAUGAAGUCCCAUGCCAUGAAGUCAAAUGCAAAAAAAAA-----AAA ..((((.....((((((((((.....))))).....--...((((........(((((......))))).(((......)))...))))..)))))...)))).........-----... ( -25.00) >consensus GAGCAUAAAACGUUUCGCUUUGGCAGGGAGCAUAAAA_AAAGGCGCACAAAAGCCGUUGUUCAAAGCGGCGUUAAUGAAGUCCCGUCCC____AGUCGAAUGCAAAAAAA_______AAA ..((((..(((((..(((((((((.....))..........(((........)))......)))))))))))).(((......))).............))))................. (-19.82 = -20.30 + 0.48)


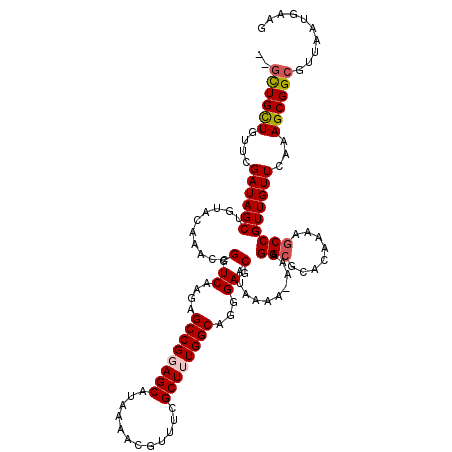
| Location | 14,932,219 – 14,932,336 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.27 |
| Mean single sequence MFE | -33.67 |
| Consensus MFE | -29.36 |
| Energy contribution | -29.76 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.689245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14932219 117 - 27905053 ---UUGCUGUUCGAUAGCUGUACAAACCGGUCAAGAGCCGGAGCAUAAAACGUUUCGCUUUGGCAGGGAGCAUAAAAAAAAGGCGCACAAAAGCCGUUGUUCAAAGCGGCGUUAAUGAAG ---..((((((.((((((........((.((((.((((.(((((.......))))))))))))).))..............(((........)))))))))...)))))).......... ( -31.00) >DroSec_CAF1 62128 119 - 1 CUGCUGUUGUUCGAUAGCUGUACAAACCGGUCAAGAGCCGCAGCAUAAAACGUUUCGCUUUGGCAGGGAGCAUAAAA-AAAGGCGCACAAAAGCCGUUGUUCAAAGCGGCGUUAAUGAAG .((((((.((((....((((.......))))...)))).))))))...(((((..(((((((((.....))......-...(((........)))......))))))))))))....... ( -35.60) >DroSim_CAF1 60824 119 - 1 CUGCUGUUGUUCGAUAGCUGUACAAACCGGUCAAGAGCCGCAGCAUAAAACGUUUCGCUUUGGCAGGGAGCAUAAAA-AAAGGCGCACAAAAGCCGUUGUUCAAAGCGGCGUUAAUGAAG .((((((.((((....((((.......))))...)))).))))))...(((((..(((((((((.....))......-...(((........)))......))))))))))))....... ( -35.60) >DroEre_CAF1 58681 116 - 1 --GCUGCUGUUCGAUAGCUGUACAAACCGGUCAAGAGCCGGAGCAUAAAACGUUUCGCUUUGGCAGGGAGCAUAAA--AAAGGCGCACAAAAGCCGUUGUUCAAAGCGGCGAUAAUGAAG --((((((....((((((........((.((((.((((.(((((.......))))))))))))).)).........--...(((........)))))))))...)))))).......... ( -34.50) >DroYak_CAF1 65688 116 - 1 --GCUGCUGUUCGAUAGCUGUACAAACCGGUCAAGAGCCGGAGCAUAAAACGCUUCGCUUUGGCGGAGAGCAUAAA--AAAGGCACACAAAAUCCGUUGUUCAAAGCGGCGAUAAUGAAG --((((((....((((((........(((((.....))))).((........((((((....))))))........--....))...........))))))...)))))).......... ( -31.65) >consensus __GCUGCUGUUCGAUAGCUGUACAAACCGGUCAAGAGCCGGAGCAUAAAACGUUUCGCUUUGGCAGGGAGCAUAAAA_AAAGGCGCACAAAAGCCGUUGUUCAAAGCGGCGUUAAUGAAG ..((((((....((((((..........(.((....((((((((............))))))))...)).)..........(((........)))))))))...)))))).......... (-29.36 = -29.76 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:26 2006