
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,930,188 – 14,930,342 |
| Length | 154 |
| Max. P | 0.999614 |

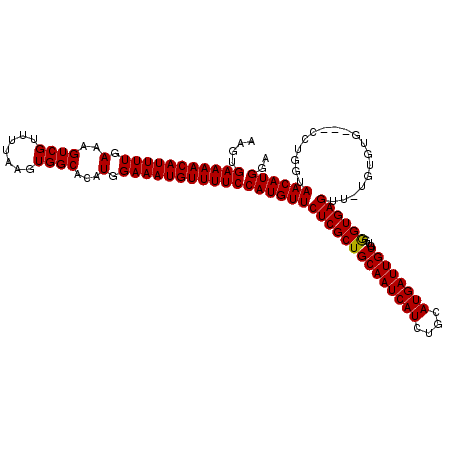
| Location | 14,930,188 – 14,930,302 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 96.36 |
| Mean single sequence MFE | -33.48 |
| Consensus MFE | -28.48 |
| Energy contribution | -28.24 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968293 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14930188 114 + 27905053 AAGUGAAAACAUUUUGAAAGUCGUUUUAAGUGGCACAUGGAAAUGUUUUCCAUGUUCUCGCUGCAAUCAUCUGCAUGAUUGUGUGGGUGAGUUUUGUGUG---CCUGGUAACAUGGA ..((....))..........((((.(((...(((((((((((.....)))).....((((((((((((((....))))))))...))))))....)))))---))...))).)))). ( -32.80) >DroSec_CAF1 60185 116 + 1 AAGUGAAAACAUUUUGAAAGUCGUUUUAAGUGGCACAUAGAAAUGUUUUCCAUGUUCUCGCUGCAAUCAUCUGCAUGAUUGUGUGGGUGAGUU-UGUGUGCCGCCUGGUAACAUGGA ..((....))............((((((.((((((((((((.(((.....)))...((((((((((((((....))))))))...))))))))-)))))))))).))).)))..... ( -34.40) >DroSim_CAF1 58879 116 + 1 AAGUGAAAACAUUUUGAAAGUCGUUUUAAGUGGCACAUGGAAAUGUUUUCCAUGUUCUCGCUGCAAUCAUCUGCAUGAUUGUGUGGGUGAGUU-UGUGUGCCGCCUGGUAACAUGGA ..((....))............((((((.(((((((((((((.....)))).....((((((((((((((....))))))))...))))))..-.))))))))).))).)))..... ( -35.30) >DroEre_CAF1 56705 113 + 1 AAGUGAAAACAUUUUGAAAGUCGUUUUAAGUGGCACAUGGAAAUGUUUUCCAUGUUCUCGCUGCAAUCAUCUGCAUGAUUGUGUGAGUGAGCU-UGUGUG---CCUGGUAACAUGGA ..((....))..........((((.(((...(((((((((((.....))))..((((((((.((((((((....))))))))))))..)))).-.)))))---))...))).)))). ( -33.10) >DroYak_CAF1 63489 113 + 1 AAGUGAAAACAUUUUGAAAGUCGUUUUAAGUGGCACAUGGAAAUGUUUUCCAUGUUCUCGCUGCAAUCAUCUGCAUGAUUGUGUGAGUGAGCU-UGUAUG---CCUGGUAACAUGGA ..((....))..........((((.(((...(((((((((((.....)))))))).((((((((((((((....))))))))...))))))..-.....)---))...))).)))). ( -31.80) >consensus AAGUGAAAACAUUUUGAAAGUCGUUUUAAGUGGCACAUGGAAAUGUUUUCCAUGUUCUCGCUGCAAUCAUCUGCAUGAUUGUGUGGGUGAGUU_UGUGUG___CCUGGUAACAUGGA ....(((((((((((.(..((((.......))))...).)))))))))))((((((((((((((((((((....))))))))...))))))..................)))))).. (-28.48 = -28.24 + -0.24)


| Location | 14,930,188 – 14,930,302 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 96.36 |
| Mean single sequence MFE | -23.82 |
| Consensus MFE | -19.40 |
| Energy contribution | -19.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14930188 114 - 27905053 UCCAUGUUACCAGG---CACACAAAACUCACCCACACAAUCAUGCAGAUGAUUGCAGCGAGAACAUGGAAAACAUUUCCAUGUGCCACUUAAAACGACUUUCAAAAUGUUUUCACUU ............((---(((......(((.(.....(((((((....)))))))..).)))...((((((.....)))))))))))............................... ( -22.60) >DroSec_CAF1 60185 116 - 1 UCCAUGUUACCAGGCGGCACACA-AACUCACCCACACAAUCAUGCAGAUGAUUGCAGCGAGAACAUGGAAAACAUUUCUAUGUGCCACUUAAAACGACUUUCAAAAUGUUUUCACUU ....((((...(((.(((((...-..(((.(.....(((((((....)))))))..).)))...((((((.....))))))))))).)))..))))..................... ( -22.90) >DroSim_CAF1 58879 116 - 1 UCCAUGUUACCAGGCGGCACACA-AACUCACCCACACAAUCAUGCAGAUGAUUGCAGCGAGAACAUGGAAAACAUUUCCAUGUGCCACUUAAAACGACUUUCAAAAUGUUUUCACUU ....((((...(((.(((((...-..(((.(.....(((((((....)))))))..).)))...((((((.....))))))))))).)))..))))..................... ( -25.20) >DroEre_CAF1 56705 113 - 1 UCCAUGUUACCAGG---CACACA-AGCUCACUCACACAAUCAUGCAGAUGAUUGCAGCGAGAACAUGGAAAACAUUUCCAUGUGCCACUUAAAACGACUUUCAAAAUGUUUUCACUU ............((---(((...-......(((.(.(((((((....)))))))..).)))...((((((.....)))))))))))............................... ( -24.20) >DroYak_CAF1 63489 113 - 1 UCCAUGUUACCAGG---CAUACA-AGCUCACUCACACAAUCAUGCAGAUGAUUGCAGCGAGAACAUGGAAAACAUUUCCAUGUGCCACUUAAAACGACUUUCAAAAUGUUUUCACUU ............((---(.....-......(((.(.(((((((....)))))))..).))).((((((((.....)))))))))))............................... ( -24.20) >consensus UCCAUGUUACCAGG___CACACA_AACUCACCCACACAAUCAUGCAGAUGAUUGCAGCGAGAACAUGGAAAACAUUUCCAUGUGCCACUUAAAACGACUUUCAAAAUGUUUUCACUU ..((((((....(........)....(((.(.....(((((((....)))))))..).)))))))))((((((((((....((.............)).....)))))))))).... (-19.40 = -19.40 + 0.00)



| Location | 14,930,225 – 14,930,342 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.12 |
| Mean single sequence MFE | -34.08 |
| Consensus MFE | -29.52 |
| Energy contribution | -29.48 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.996704 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14930225 117 + 27905053 UGGAAAUGUUUUCCAUGUUCUCGCUGCAAUCAUCUGCAUGAUUGUGUGGGUGAGUUUUGUGUG---CCUGGUAACAUGGAGUAAUUUUCCAUUUCUAAAAUGUGCAAAAUGCGUUGAUUU ((((((....(((((((((((.((((((((((((..(((....)))..)))))...))))).)---)..)).)))))))))....)))))).......(((((.......)))))..... ( -34.40) >DroSec_CAF1 60222 119 + 1 UAGAAAUGUUUUCCAUGUUCUCGCUGCAAUCAUCUGCAUGAUUGUGUGGGUGAGUU-UGUGUGCCGCCUGGUAACAUGGAGUAAUUUUCCAUUUCUAAAAUGUGCAAAAUGCGUUGAUUU ((((((((..(((((((((((.((.(((.(((((..(((....)))..)))))...-....))).))..)).)))))))))........)))))))).(((((.......)))))..... ( -33.90) >DroSim_CAF1 58916 119 + 1 UGGAAAUGUUUUCCAUGUUCUCGCUGCAAUCAUCUGCAUGAUUGUGUGGGUGAGUU-UGUGUGCCGCCUGGUAACAUGGAGUAAUUUUCCAUUUCUAAAAUGUGCAAAAUGCGUUGAUUU ((((((....(((((((((((.((.(((.(((((..(((....)))..)))))...-....))).))..)).)))))))))....)))))).......(((((.......)))))..... ( -35.70) >DroEre_CAF1 56742 116 + 1 UGGAAAUGUUUUCCAUGUUCUCGCUGCAAUCAUCUGCAUGAUUGUGUGAGUGAGCU-UGUGUG---CCUGGUAACAUGGAGUAAUUUUCCAUUUCUAAAAUGUGCAAAAUGCGUUGAUUU ((((((....((((((((((((((.((((((((....)))))))))))))...((.-.....)---).....)))))))))....)))))).......(((((.......)))))..... ( -33.20) >DroYak_CAF1 63526 116 + 1 UGGAAAUGUUUUCCAUGUUCUCGCUGCAAUCAUCUGCAUGAUUGUGUGAGUGAGCU-UGUAUG---CCUGGUAACAUGGAGUAAUUUUCCAUUUCUAAAAUGUGCAAAAUGCGUUGACUU ((((((....((((((((((((((.((((((((....)))))))))))))...((.-.....)---).....)))))))))....)))))).......(((((.......)))))..... ( -33.20) >consensus UGGAAAUGUUUUCCAUGUUCUCGCUGCAAUCAUCUGCAUGAUUGUGUGGGUGAGUU_UGUGUG___CCUGGUAACAUGGAGUAAUUUUCCAUUUCUAAAAUGUGCAAAAUGCGUUGAUUU ((((((....(((((((((((((((((((((((....))))))))...))))))..................)))))))))....)))))).......(((((.......)))))..... (-29.52 = -29.48 + -0.04)

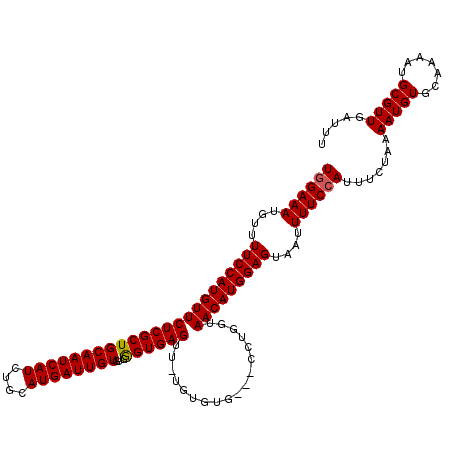

| Location | 14,930,225 – 14,930,342 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.12 |
| Mean single sequence MFE | -25.76 |
| Consensus MFE | -23.08 |
| Energy contribution | -22.92 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.79 |
| SVM RNA-class probability | 0.999614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14930225 117 - 27905053 AAAUCAACGCAUUUUGCACAUUUUAGAAAUGGAAAAUUACUCCAUGUUACCAGG---CACACAAAACUCACCCACACAAUCAUGCAGAUGAUUGCAGCGAGAACAUGGAAAACAUUUCCA ........((.....))............((((((.....((((((((....(.---....)....(((.(.....(((((((....)))))))..).))))))))))).....)))))) ( -23.50) >DroSec_CAF1 60222 119 - 1 AAAUCAACGCAUUUUGCACAUUUUAGAAAUGGAAAAUUACUCCAUGUUACCAGGCGGCACACA-AACUCACCCACACAAUCAUGCAGAUGAUUGCAGCGAGAACAUGGAAAACAUUUCUA ........((.....))......((((((((.........((((((((.((....))......-..(((.(.....(((((((....)))))))..).)))))))))))...)))))))) ( -24.60) >DroSim_CAF1 58916 119 - 1 AAAUCAACGCAUUUUGCACAUUUUAGAAAUGGAAAAUUACUCCAUGUUACCAGGCGGCACACA-AACUCACCCACACAAUCAUGCAGAUGAUUGCAGCGAGAACAUGGAAAACAUUUCCA ........((.....))............((((((.....((((((((.((....))......-..(((.(.....(((((((....)))))))..).))))))))))).....)))))) ( -24.90) >DroEre_CAF1 56742 116 - 1 AAAUCAACGCAUUUUGCACAUUUUAGAAAUGGAAAAUUACUCCAUGUUACCAGG---CACACA-AGCUCACUCACACAAUCAUGCAGAUGAUUGCAGCGAGAACAUGGAAAACAUUUCCA ........((.....))............((((((.....((((((((....((---(.....-.)))..(((.(.(((((((....)))))))..).))))))))))).....)))))) ( -27.90) >DroYak_CAF1 63526 116 - 1 AAGUCAACGCAUUUUGCACAUUUUAGAAAUGGAAAAUUACUCCAUGUUACCAGG---CAUACA-AGCUCACUCACACAAUCAUGCAGAUGAUUGCAGCGAGAACAUGGAAAACAUUUCCA ........((.....))............((((((.....((((((((....((---(.....-.)))..(((.(.(((((((....)))))))..).))))))))))).....)))))) ( -27.90) >consensus AAAUCAACGCAUUUUGCACAUUUUAGAAAUGGAAAAUUACUCCAUGUUACCAGG___CACACA_AACUCACCCACACAAUCAUGCAGAUGAUUGCAGCGAGAACAUGGAAAACAUUUCCA ........((.....))............((((((.....((((((((....(........)....(((.(.....(((((((....)))))))..).))))))))))).....)))))) (-23.08 = -22.92 + -0.16)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:20 2006