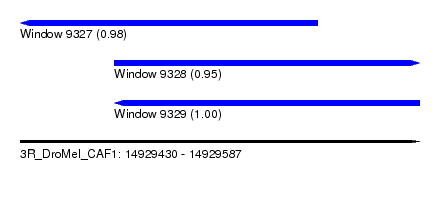
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,929,430 – 14,929,587 |
| Length | 157 |
| Max. P | 0.995861 |

| Location | 14,929,430 – 14,929,547 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.73 |
| Mean single sequence MFE | -30.61 |
| Consensus MFE | -26.08 |
| Energy contribution | -25.80 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14929430 117 - 27905053 CGGCAACAAUUGCCGCCAAUCGAAGCUGUUUUUCUUAUUGAUUAAGCGCGCUUUUAUUGGCGUUUGCCAUCGAUGUCCAGU---CCAUCACCAUCACUCUCUCCUCGCGCGGCUCUCUUU .((((.....))))(((.......(((.....((.....))...)))((((......((((....))))..((((......---.)))).................)))))))....... ( -29.80) >DroSec_CAF1 59410 120 - 1 CGGCAACAAUUGCCGCCAAUCGAAGCUGUUUUUCUUAUUGAUUAAGCGCGCUUUUAUUGGCGUUUGCCAGCGAUGUCCAGUUCACCAUCACCAUCACUCUCUCCUCGCGCGGCUCUCUUU .((((.....))))((((((.((((((((((.((.....))..))))).))))).))))))....(((.((((.(...(((..............)))....).))))..)))....... ( -33.34) >DroSim_CAF1 58109 120 - 1 CGGCAACAAUUGCCGCCAAUCGAAGCUGUUUUUCUUAUUGAUUAAGCGCGCUUUUAUUGGCGUUUGCCAUCGAUGUCCAGUUCACCAUCACCAUCACUCUCUCCUCGCGCGGCUCUCUUU .((((.....))))(((.......(((.....((.....))...)))((((......((((....))))..((((..........)))).................)))))))....... ( -29.20) >DroEre_CAF1 55968 111 - 1 CGGCAACAAUUGUUGCCAAUCGAAGCUGUUUUUCUUAUUGAUUAAGCGCGCUUUUAUUGGCGUUUGCCAUCGAUGUCCAGUG---------CAUCACUCUCUCCUCGGUCGGCUGUCUUU .((((.........((((((.((((((((((.((.....))..))))).))))).))))))....((((((((.(...((((---------...))))....).))))).)))))))... ( -32.50) >DroYak_CAF1 62769 111 - 1 CGGCUACAAUUGUUGCCAAUCGAAGCUGUUUUUCGAAUUGAUUAAGCGUGCUUUUAUUGGCGUUUGCCAUCGAUGUCCAGUG---------CAUCACUCUCUCCUCGGUCGGCUCUCUUU ..............((((((.((((((((((.((.....))..))))).))))).))))))....((((((((.(...((((---------...))))....).))))).)))....... ( -28.20) >consensus CGGCAACAAUUGCCGCCAAUCGAAGCUGUUUUUCUUAUUGAUUAAGCGCGCUUUUAUUGGCGUUUGCCAUCGAUGUCCAGUG__CCAUCACCAUCACUCUCUCCUCGCGCGGCUCUCUUU .((((.....))))((((((.((((((((((.((.....))..))))).))))).))))))....(((..(((.(...(((..............)))....).)))...)))....... (-26.08 = -25.80 + -0.28)


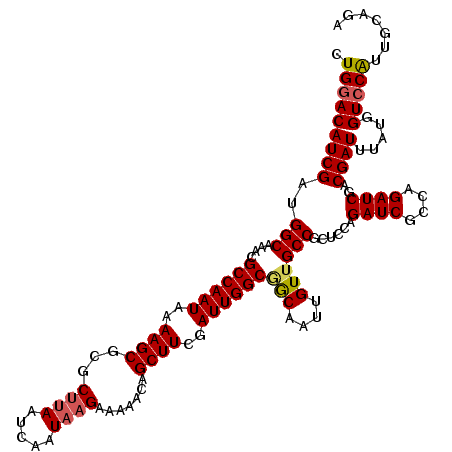
| Location | 14,929,467 – 14,929,587 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -34.82 |
| Consensus MFE | -29.72 |
| Energy contribution | -30.00 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.952911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14929467 120 + 27905053 CUGGACAUCGAUGGCAAACGCCAAUAAAAGCGCGCUUAAUCAAUAAGAAAAACAGCUUCGAUUGGCGGCAAUUGUUGCCGCUCCAGAUCGCCAGAUCGACGAUUUAUGGUCCAUUGCAGA .(((((((((..(((....((((((..((((...((((.....)))).......))))..))))))((((.....)))))))...((((....))))..)))).....)))))....... ( -35.80) >DroSec_CAF1 59450 120 + 1 CUGGACAUCGCUGGCAAACGCCAAUAAAAGCGCGCUUAAUCAAUAAGAAAAACAGCUUCGAUUGGCGGCAAUUGUUGCCGCUCCAGAUCGCCAGAUCGACGAUUUAUGGUCCAUUGCAGA .(((((...((.(((((.(((((((..((((...((((.....)))).......))))..))))))).......)))))))....((((....))))...........)))))....... ( -36.10) >DroSim_CAF1 58149 120 + 1 CUGGACAUCGAUGGCAAACGCCAAUAAAAGCGCGCUUAAUCAAUAAGAAAAACAGCUUCGAUUGGCGGCAAUUGUUGCCGCUCCAGAUCGCCAGAUCGACGAUUUAUGGUCCAUUGCAGA .(((((((((..(((....((((((..((((...((((.....)))).......))))..))))))((((.....)))))))...((((....))))..)))).....)))))....... ( -35.80) >DroEre_CAF1 55999 119 + 1 CUGGACAUCGAUGGCAAACGCCAAUAAAAGCGCGCUUAAUCAAUAAGAAAAACAGCUUCGAUUGGCAACAAUUGUUGCCGCUCCAGAUCGC-AGAUCGACGAUUUAUGGUCCGUGGUAGA (((((((((((((((....)))).....(((...((((.....)))).......)))))))).((((((....))))))).)))))(((((-.(((((........))))).)))))... ( -35.80) >DroYak_CAF1 62800 119 + 1 CUGGACAUCGAUGGCAAACGCCAAUAAAAGCACGCUUAAUCAAUUCGAAAAACAGCUUCGAUUGGCAACAAUUGUAGCCGCUCCAGAUCGC-AGAUCGACGAUUUAUGGUGCAUGGUAGU .......((((((((....))))....(((....))).......))))......(((.((((((....)))))).))).((.((((((((.-.......)))))..))).))........ ( -30.60) >consensus CUGGACAUCGAUGGCAAACGCCAAUAAAAGCGCGCUUAAUCAAUAAGAAAAACAGCUUCGAUUGGCGGCAAUUGUUGCCGCUCCAGAUCGCCAGAUCGACGAUUUAUGGUCCAUUGCAGA .(((((((((..(((....((((((..((((...((((.....)))).......))))..))))))(((....))))))......((((....))))..)))).....)))))....... (-29.72 = -30.00 + 0.28)


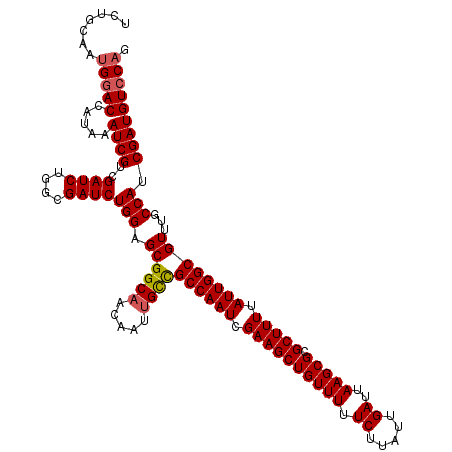
| Location | 14,929,467 – 14,929,587 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -38.22 |
| Consensus MFE | -33.90 |
| Energy contribution | -34.02 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.995861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14929467 120 - 27905053 UCUGCAAUGGACCAUAAAUCGUCGAUCUGGCGAUCUGGAGCGGCAACAAUUGCCGCCAAUCGAAGCUGUUUUUCUUAUUGAUUAAGCGCGCUUUUAUUGGCGUUUGCCAUCGAUGUCCAG .......(((((........(((((..((((((.(......((((.....))))((((((.((((((((((.((.....))..))))).))))).))))))).)))))))))))))))). ( -41.70) >DroSec_CAF1 59450 120 - 1 UCUGCAAUGGACCAUAAAUCGUCGAUCUGGCGAUCUGGAGCGGCAACAAUUGCCGCCAAUCGAAGCUGUUUUUCUUAUUGAUUAAGCGCGCUUUUAUUGGCGUUUGCCAGCGAUGUCCAG .......(((((........((((..(((((((.(......((((.....))))((((((.((((((((((.((.....))..))))).))))).))))))).)))))))))))))))). ( -42.40) >DroSim_CAF1 58149 120 - 1 UCUGCAAUGGACCAUAAAUCGUCGAUCUGGCGAUCUGGAGCGGCAACAAUUGCCGCCAAUCGAAGCUGUUUUUCUUAUUGAUUAAGCGCGCUUUUAUUGGCGUUUGCCAUCGAUGUCCAG .......(((((........(((((..((((((.(......((((.....))))((((((.((((((((((.((.....))..))))).))))).))))))).)))))))))))))))). ( -41.70) >DroEre_CAF1 55999 119 - 1 UCUACCACGGACCAUAAAUCGUCGAUCU-GCGAUCUGGAGCGGCAACAAUUGUUGCCAAUCGAAGCUGUUUUUCUUAUUGAUUAAGCGCGCUUUUAUUGGCGUUUGCCAUCGAUGUCCAG ........((((........((((((..-((........))((((((....)))((((((.((((((((((.((.....))..))))).))))).))))))....))))))))))))).. ( -35.30) >DroYak_CAF1 62800 119 - 1 ACUACCAUGCACCAUAAAUCGUCGAUCU-GCGAUCUGGAGCGGCUACAAUUGUUGCCAAUCGAAGCUGUUUUUCGAAUUGAUUAAGCGUGCUUUUAUUGGCGUUUGCCAUCGAUGUCCAG .....(((((.(((...(((((......-))))).))).))(((.((....)).((((((.((((((((((.((.....))..))))).))))).))))))....)))....)))..... ( -30.00) >consensus UCUGCAAUGGACCAUAAAUCGUCGAUCUGGCGAUCUGGAGCGGCAACAAUUGCCGCCAAUCGAAGCUGUUUUUCUUAUUGAUUAAGCGCGCUUUUAUUGGCGUUUGCCAUCGAUGUCCAG .......(((((.....((((..((((....))))(((.((((((.....))))((((((.((((((((((.((.....))..))))).))))).))))))))...))).))))))))). (-33.90 = -34.02 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:16 2006