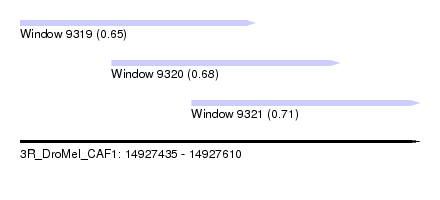
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,927,435 – 14,927,610 |
| Length | 175 |
| Max. P | 0.709269 |

| Location | 14,927,435 – 14,927,538 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.35 |
| Mean single sequence MFE | -30.82 |
| Consensus MFE | -15.20 |
| Energy contribution | -16.68 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14927435 103 + 27905053 AUCAGCUGGGGGAGCUGCACUUUUCCGACAGCCCCUGCAAUUUAUGCCCCCG-AAAAUGG-GGGA-AUAGCUC--UCAAGCACCAGAAGAUUCCCUCAAAAAAG---UGAA--------- (((..(((((((.(((((........).))))))).......(((.(((((.-.....))-))).-)))(((.--...))).))))..))).............---....--------- ( -30.10) >DroSec_CAF1 57441 105 + 1 AUCAGCUGGGGGAGCUGCACUUUUCCGGCAGCCACAGCAAUUUAUGCCUCCGGAAAAUGGGGGGA-AAAGUUC--UCAAGCACCAGAAGAUACCCUCAAAAAAG---UGGA--------- ....(((((..(((.....)))..)))))..((((.(((.....)))..........((((((..-....(((--(........))))....)))))).....)---))).--------- ( -31.60) >DroSim_CAF1 56113 108 + 1 AUCAGCUGGGGGAGCUGCACUUUUCCGGCAGCCACAGCAAUUUAUGCCCCCGGAAAAUGGGGGGA-AAAGUUC--UCAAGCACCAGAAGAUACCCUCAAAAAAGAAAUAGA--------- (((.(((((..(((.....)))..))))).((...((.(((((.(.((((((.....)))))).)-.))))))--)...)).......)))....................--------- ( -31.80) >DroEre_CAF1 53972 98 + 1 AUCAGCUGGGGGAGCUGCACUUUUCCGGCAC-----------UUUUCCCCCG-AAAAUGCG--UACUAAGUUCUCUCAAGUACCAGAAAAACCCCUGAAAAACA---GGCAGCGG----- ....(((((..(((.....)))..)))))..-----------.......(((-....((((--((((.((....))..)))))(((........))).......---.))).)))----- ( -26.10) >DroYak_CAF1 60792 111 + 1 AUCAACUGGGGGAGCUGCACUUUUCCGGCAGCCCCUGCAAUUUAUGCCCCCG-AAAAUGGG--AA-AAAGUUC--UCAAGUACCAGAUGAUCCCCUGAAAAACG---GGCAUAAAAACAA ........((((.(((((.(......))))))))))....(((((((((...-....((((--((-....)))--))).....(((........)))......)---))))))))..... ( -34.50) >consensus AUCAGCUGGGGGAGCUGCACUUUUCCGGCAGCCACAGCAAUUUAUGCCCCCG_AAAAUGGGGGGA_AAAGUUC__UCAAGCACCAGAAGAUACCCUCAAAAAAG___UGCA_________ .((.(((((..(((.....)))..))))).................((((((.....))))))......................))................................. (-15.20 = -16.68 + 1.48)


| Location | 14,927,475 – 14,927,575 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.89 |
| Mean single sequence MFE | -19.36 |
| Consensus MFE | -3.10 |
| Energy contribution | -4.18 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.16 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14927475 100 + 27905053 UUUAUGCCCCCG-AAAAUGG-GGGA-AUAGCUC--UCAAGCACCAGAAGAUUCCCUCAAAAAAG---UGAA----------CAGAGA--AAUUCCAAAUUUUUCAUUUCUAUUUUUAAAA ............-....(((-((((-((...((--(........)))..))))))))).(((((---((((----------..((((--(((.....)))))))..))).)))))).... ( -20.10) >DroSec_CAF1 57481 101 + 1 UUUAUGCCUCCGGAAAAUGGGGGGA-AAAGUUC--UCAAGCACCAGAAGAUACCCUCAAAAAAG---UGGA----------CAGAGA--AAUUCCAAUUUUCCCAUUUCUAUUUUUA-AA ............(((((((((((((-(((.(((--((......(....)...((((......))---.)).----------..))))--).......)))))))...))))))))).-.. ( -17.50) >DroSim_CAF1 56153 107 + 1 UUUAUGCCCCCGGAAAAUGGGGGGA-AAAGUUC--UCAAGCACCAGAAGAUACCCUCAAAAAAGAAAUAGA----------CAGAGAAAAAUUUCAAAUUUCCCAUUUCUAUUUUUAAAA ....(.((((((.....)))))).)-....(((--(........))))..............(((((((((----------..(.((((.........)))).)...))))))))).... ( -17.80) >DroEre_CAF1 54003 104 + 1 --UUUUCCCCCG-AAAAUGCG--UACUAAGUUCUCUCAAGUACCAGAAAAACCCCUGAAAAACA---GGCAGCGG------CGGAGA--AAUUCCAAAUUUUCCAUUUCUAUUUUUAAAA --((((((.(((-....((((--((((.((....))..)))))(((........))).......---.))).)))------.)))))--).............................. ( -19.60) >DroYak_CAF1 60832 109 + 1 UUUAUGCCCCCG-AAAAUGGG--AA-AAAGUUC--UCAAGUACCAGAUGAUCCCCUGAAAAACG---GGCAUAAAAACAAAUUUAAA--GAUUCCAAAUUUCCCAUUUCUAUUUUUAAAA (((((((((...-....((((--((-....)))--))).....(((........)))......)---))))))))......((((((--(((...((((.....))))..))))))))). ( -21.80) >consensus UUUAUGCCCCCG_AAAAUGGGGGGA_AAAGUUC__UCAAGCACCAGAAGAUACCCUCAAAAAAG___UGCA__________CAGAGA__AAUUCCAAAUUUCCCAUUUCUAUUUUUAAAA ......((((((.....))))))................................................................................................. ( -3.10 = -4.18 + 1.08)



| Location | 14,927,510 – 14,927,610 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.45 |
| Mean single sequence MFE | -20.51 |
| Consensus MFE | -1.40 |
| Energy contribution | -1.64 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.07 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14927510 100 + 27905053 CACCAGAAGAUUCCCUCAAAAAAG---UGAA----------CAGAGA--AAUUCCAAAUUUUUCAUUUCUAUUUUUAAAAUGGCAGCGG-----AACAACGUUGCCUAGUUUUUCAAAUU .....(((((.........(((((---((((----------..((((--(((.....)))))))..))).)))))).....(((((((.-----.....)))))))....)))))..... ( -21.70) >DroSec_CAF1 57518 99 + 1 CACCAGAAGAUACCCUCAAAAAAG---UGGA----------CAGAGA--AAUUCCAAUUUUCCCAUUUCUAUUUUUA-AAUAGCAGAGG-----AGCAACAUUGCCCAGUUUUUUAAAUU ....(((((((..((((....(((---(((.----------..((((--(.......)))))))))))(((((....-)))))..))))-----.(((....)))...)))))))..... ( -15.40) >DroSim_CAF1 56190 107 + 1 CACCAGAAGAUACCCUCAAAAAAGAAAUAGA----------CAGAGAAAAAUUUCAAAUUUCCCAUUUCUAUUUUUAAAAUAGCAGAGGA---GAGCAACGUUGCCCAGUUUUUCAAAUU .....(((((.((((((.....(((((((((----------..(.((((.........)))).)...))))))))).........)))).---(.(((....))).).)))))))..... ( -13.64) >DroEre_CAF1 54038 104 + 1 UACCAGAAAAACCCCUGAAAAACA---GGCAGCGG------CGGAGA--AAUUCCAAAUUUUCCAUUUCUAUUUUUAAAAGUCCAGGGGA-----GCAACGUUGCCUAGUUUUUCAAAUU ...............(((((((((---((((((((------((((((--(........)))))).................(((...)))-----))..)))))))).)))))))).... ( -29.00) >DroYak_CAF1 60866 115 + 1 UACCAGAUGAUCCCCUGAAAAACG---GGCAUAAAAACAAAUUUAAA--GAUUCCAAAUUUCCCAUUUCUAUUUUUAAAAGUCCAGGGGAGCGGAGCGACGUUGCCUGGUUUUUCAAAUU .(((((....(((((((......(---..........)...((((((--(((...((((.....))))..)))))))))....)))))))(((......)))...))))).......... ( -22.80) >consensus CACCAGAAGAUACCCUCAAAAAAG___UGCA__________CAGAGA__AAUUCCAAAUUUCCCAUUUCUAUUUUUAAAAUAGCAGAGGA____AGCAACGUUGCCUAGUUUUUCAAAUU .....(((((................................(((((..((........))....)))))............((((.(...........).)))).....)))))..... ( -1.40 = -1.64 + 0.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:08 2006