
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,927,047 – 14,927,395 |
| Length | 348 |
| Max. P | 0.999136 |

| Location | 14,927,047 – 14,927,166 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.52 |
| Mean single sequence MFE | -19.63 |
| Consensus MFE | -17.89 |
| Energy contribution | -18.69 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14927047 119 + 27905053 -UUUCAACAAGCAUUCCACGCAUGGCCAUCAAAGUAAAUUACUUAAUUUAUGCCUCAAAAUGUGUCGUCGAGCGGGCGGCACUUUCUACCUCUGCCACCCAACAAAACAAAAAAAAAAUA -.........((...(.(((((((((.......(((((((....)))))))))).....)))))).)....))(((.((((...........)))).))).................... ( -21.41) >DroSec_CAF1 57055 112 + 1 -UUUCAACAAGCAUUCCACGCAUGGCCAUCAAAGUAAAUUACUUAAUUUAUGCCUCAAAAUGUGUCGUCGAGCGGGCGGCACUUUCUAUCUCUGCCACCCAACAA-------CAAAAAUA -.........((...(.(((((((((.......(((((((....)))))))))).....)))))).)....))(((.((((...........)))).))).....-------........ ( -21.41) >DroSim_CAF1 55729 112 + 1 -UUUCAACAAGCAUUCCACGCAUGGCCAUCAAAGUAAAUUACUUAAUUUAUGCCUCAAAAUGUGUCGUCGAGCGGGCGGCACUUUCUACCUCUGCCACCCAACAA-------CAAAAAUA -.........((...(.(((((((((.......(((((((....)))))))))).....)))))).)....))(((.((((...........)))).))).....-------........ ( -21.41) >DroEre_CAF1 53623 102 + 1 -UUUCAACAAGCAUUCCACGCAUGGCCAUCAAAGUAAAUUACUUAAUUUAUGCCUCAAAAUGUGUCGUCGAGCGGGCGGCACUUUCUA------CCACCCAAC-----------AAAAUA -.........((.......))..(((.......(((((((....)))))))))).......((((((((.....))))))))......------.........-----------...... ( -17.01) >DroYak_CAF1 60440 105 + 1 UUUUCAACAAGCAUUCCACGCAUGACCAUCAAAGUAAAUUACUUAAUUUAUGCCUCAAAAUGUGUCGUCGAGCGGGCGGCACUUUCUA------CCACCCAAC---------CAAAAAUA ...................((((((......(((((...)))))...))))))........((((((((.....))))))))......------.........---------........ ( -16.90) >consensus _UUUCAACAAGCAUUCCACGCAUGGCCAUCAAAGUAAAUUACUUAAUUUAUGCCUCAAAAUGUGUCGUCGAGCGGGCGGCACUUUCUA_CUCUGCCACCCAACAA_______CAAAAAUA ..........((...(.((((((..........(((((((....)))))))........)))))).)....))(((.((((...........)))).))).................... (-17.89 = -18.69 + 0.80)


| Location | 14,927,126 – 14,927,246 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.59 |
| Mean single sequence MFE | -26.12 |
| Consensus MFE | -21.00 |
| Energy contribution | -23.00 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14927126 120 + 27905053 ACUUUCUACCUCUGCCACCCAACAAAACAAAAAAAAAAUACAAAGCGGUUCUUGCGUAAGCGAUUUUAAACGCAUGCAAAACGCGUGCAGCGAAAGCAGGGCAAACUUUUGUGUGUACUU .....................................((((((((..((((((((....))))........((((((.....)))))).((....)).))))....))))))))...... ( -27.30) >DroSec_CAF1 57134 109 + 1 ACUUUCUAUCUCUGCCACCCAACAA-------CAAAAAUACAAA----UUCUUGCGUAAGCGAUUUUAAACGCAUGCAAAACGCGUGCAGCAAAAGCAGGGCAAACUUUUGUGUGUACUU .........((((((..........-------............----...((((....))))........((((((.....)))))).......))))))................... ( -22.60) >DroSim_CAF1 55808 113 + 1 ACUUUCUACCUCUGCCACCCAACAA-------CAAAAAUACAAAGCGGUUCUUGCGUAAGCGACUUUAAACGCAUGCAAAACGCGUGCAGCGAAAGCAGGGCAAACUUUUGUGUGUACUU .........................-------.....((((((((..((((((((....))))........((((((.....)))))).((....)).))))....))))))))...... ( -27.20) >DroEre_CAF1 53702 101 + 1 ACUUUCUA------CCACCCAAC-----------AAAAUACAAAGCGGUUCUUGCGUAAGCGACUUUAAACGCAUGCAAAACGCGUGCAGCGAAAGCAGGGCAAACUCUUGU--GUACUU ........------.........-----------...((((((((..((((((((....))))........((((((.....)))))).((....)).))))...)).))))--)).... ( -26.30) >DroYak_CAF1 60520 103 + 1 ACUUUCUA------CCACCCAAC---------CAAAAAUACAAAGCGGUUCUUGCGUAAGCGACUUUAAACGCAUGCAAAACGCGUGCAGCGAAAGCAGGGCAAACUUUUGU--GUACUU ........------.........---------.....((((((((..((((((((....))))........((((((.....)))))).((....)).))))....))))))--)).... ( -27.20) >consensus ACUUUCUA_CUCUGCCACCCAACAA_______CAAAAAUACAAAGCGGUUCUUGCGUAAGCGACUUUAAACGCAUGCAAAACGCGUGCAGCGAAAGCAGGGCAAACUUUUGUGUGUACUU .................(((...............................((((....))))........((((((.....)))))).((....)).)))................... (-21.00 = -23.00 + 2.00)



| Location | 14,927,166 – 14,927,281 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.34 |
| Mean single sequence MFE | -29.75 |
| Consensus MFE | -22.00 |
| Energy contribution | -22.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14927166 115 + 27905053 CAAAGCGGUUCUUGCGUAAGCGAUUUUAAACGCAUGCAAAACGCGUGCAGCGAAAGCAGGGCAAACUUUUGUGUGUACUUAUGUAUGAAUGUAUGUAU----AU-UGUUACAUCAGAUUU .......((((.((((((((...........((((((.....)))))).((....))...(((.((....)).))).)))))))).))))(.(((((.----..-...))))))...... ( -30.00) >DroSim_CAF1 55841 116 + 1 CAAAGCGGUUCUUGCGUAAGCGACUUUAAACGCAUGCAAAACGCGUGCAGCGAAAGCAGGGCAAACUUUUGUGUGUACUUAUGUAUG----UAUGUAUAUACAUCAGUUACAUCAGAUUU .......((((((((....))))........((((((.....)))))).((....)).)))).((((..(((((((((.(((....)----)).)))))))))..))))........... ( -35.20) >DroEre_CAF1 53725 95 + 1 CAAAGCGGUUCUUGCGUAAGCGACUUUAAACGCAUGCAAAACGCGUGCAGCGAAAGCAGGGCAAACUCUUGU--GUACUUAUAU---------------------AGUUACAUCAGAU-- .......((((((((....))))........((((((.....)))))).((....)).))))....(((.((--(((((.....---------------------)).))))).))).-- ( -27.60) >DroYak_CAF1 60545 95 + 1 CAAAGCGGUUCUUGCGUAAGCGACUUUAAACGCAUGCAAAACGCGUGCAGCGAAAGCAGGGCAAACUUUUGU--GUACUUAUAU---------------------GGUUACAUCAGAU-- .......((((((((....))))........((((((.....)))))).((....)).))))...((..(((--(.(((.....---------------------)))))))..))..-- ( -26.20) >consensus CAAAGCGGUUCUUGCGUAAGCGACUUUAAACGCAUGCAAAACGCGUGCAGCGAAAGCAGGGCAAACUUUUGU__GUACUUAUAU_____________________AGUUACAUCAGAU__ ....(((....((((((..(((........)))))))))..)))((((.((....))...((((....))))..)))).......................................... (-22.00 = -22.00 + 0.00)



| Location | 14,927,206 – 14,927,321 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.79 |
| Mean single sequence MFE | -32.08 |
| Consensus MFE | -26.82 |
| Energy contribution | -26.82 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991170 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
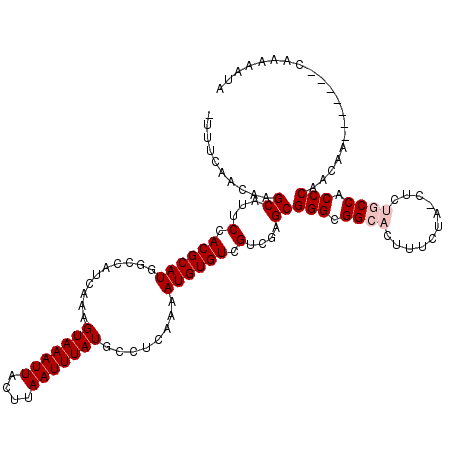
>3R_DroMel_CAF1 14927206 115 + 27905053 ACGCGUGCAGCGAAAGCAGGGCAAACUUUUGUGUGUACUUAUGUAUGAAUGUAUGUAU----AU-UGUUACAUCAGAUUUUCCCAGUGACACGGGGUCACUGGAAAUUAACUGAUAAAAG ..(((((((((....))((..((.((....)).))..))..........)))))))..----..-......(((((..(((((.((((((.....)))))))))))....)))))..... ( -33.00) >DroSim_CAF1 55881 116 + 1 ACGCGUGCAGCGAAAGCAGGGCAAACUUUUGUGUGUACUUAUGUAUG----UAUGUAUAUACAUCAGUUACAUCAGAUUUUCCCAGUGACACGGGGUCACUGGAAAUUAACUGAUAAAAG ....((.(.((....)).).)).((((..(((((((((.(((....)----)).)))))))))..))))..(((((..(((((.((((((.....)))))))))))....)))))..... ( -38.50) >DroEre_CAF1 53765 95 + 1 ACGCGUGCAGCGAAAGCAGGGCAAACUCUUGU--GUACUUAUAU---------------------AGUUACAUCAGAU--UCCCAGUGACACGGGGUCACUGGAAAUUAACUGAUAAAAG ....(((((((....))((((....))))..)--))))......---------------------......(((((((--(.((((((((.....)))))))).)))...)))))..... ( -28.80) >DroYak_CAF1 60585 95 + 1 ACGCGUGCAGCGAAAGCAGGGCAAACUUUUGU--GUACUUAUAU---------------------GGUUACAUCAGAU--UCCCAGUGACACGGGGUCACUGGAAAUUAACUGAUAAAAG ....((.(.((....)).).))...(((((((--(.(((.....---------------------))))))(((((((--(.((((((((.....)))))))).)))...)))))))))) ( -28.00) >consensus ACGCGUGCAGCGAAAGCAGGGCAAACUUUUGU__GUACUUAUAU_____________________AGUUACAUCAGAU__UCCCAGUGACACGGGGUCACUGGAAAUUAACUGAUAAAAG ....((((.((....))...((((....))))..)))).................................(((((......((((((((.....)))))))).......)))))..... (-26.82 = -26.82 + 0.00)


| Location | 14,927,206 – 14,927,321 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.79 |
| Mean single sequence MFE | -28.45 |
| Consensus MFE | -26.25 |
| Energy contribution | -26.25 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.85 |
| SVM RNA-class probability | 0.997405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14927206 115 - 27905053 CUUUUAUCAGUUAAUUUCCAGUGACCCCGUGUCACUGGGAAAAUCUGAUGUAACA-AU----AUACAUACAUUCAUACAUAAGUACACACAAAAGUUUGCCCUGCUUUCGCUGCACGCGU ....((((((....(((((((((((.....)))))))))))...)))))).....-..----................................((.(((...((....)).))).)).. ( -27.00) >DroSim_CAF1 55881 116 - 1 CUUUUAUCAGUUAAUUUCCAGUGACCCCGUGUCACUGGGAAAAUCUGAUGUAACUGAUGUAUAUACAUA----CAUACAUAAGUACACACAAAAGUUUGCCCUGCUUUCGCUGCACGCGU ....((((((....(((((((((((.....)))))))))))...))))))..(((.((((((.......----.)))))).)))..........((.(((...((....)).))).)).. ( -32.60) >DroEre_CAF1 53765 95 - 1 CUUUUAUCAGUUAAUUUCCAGUGACCCCGUGUCACUGGGA--AUCUGAUGUAACU---------------------AUAUAAGUAC--ACAAGAGUUUGCCCUGCUUUCGCUGCACGCGU ....((((((...((((((((((((.....))))))))))--)))))))).....---------------------..........--......((.(((...((....)).))).)).. ( -27.10) >DroYak_CAF1 60585 95 - 1 CUUUUAUCAGUUAAUUUCCAGUGACCCCGUGUCACUGGGA--AUCUGAUGUAACC---------------------AUAUAAGUAC--ACAAAAGUUUGCCCUGCUUUCGCUGCACGCGU ....((((((...((((((((((((.....))))))))))--)))))))).....---------------------..........--......((.(((...((....)).))).)).. ( -27.10) >consensus CUUUUAUCAGUUAAUUUCCAGUGACCCCGUGUCACUGGGA__AUCUGAUGUAACU_____________________ACAUAAGUAC__ACAAAAGUUUGCCCUGCUUUCGCUGCACGCGU ....((((((.....((((((((((.....))))))))))....))))))............................................((.(((...((....)).))).)).. (-26.25 = -26.25 + -0.00)


| Location | 14,927,246 – 14,927,361 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.68 |
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -23.72 |
| Energy contribution | -24.84 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14927246 115 + 27905053 AUGUAUGAAUGUAUGUAU----AU-UGUUACAUCAGAUUUUCCCAGUGACACGGGGUCACUGGAAAUUAACUGAUAAAAGCUGGACGAACAGCUCCAUCUUUGGACUUUGCUCCCCCCGG ..(((..(((((....))----))-)..)))(((((..(((((.((((((.....)))))))))))....)))))....((((......))))((((....))))............... ( -29.70) >DroSim_CAF1 55921 114 + 1 AUGUAUG----UAUGUAUAUACAUCAGUUACAUCAGAUUUUCCCAGUGACACGGGGUCACUGGAAAUUAACUGAUAAAAGCGGGGCGAGCAGCUCCAUUUUUGGACUUUGCUCC--ACGG (((((((----(....))))))))..(((..(((((..(((((.((((((.....)))))))))))....)))))...)))(((((((..((.((((....)))))))))))))--.... ( -38.30) >DroEre_CAF1 53803 95 + 1 AUAU---------------------AGUUACAUCAGAU--UCCCAGUGACACGGGGUCACUGGAAAUUAACUGAUAAAAGCUGGGCGAGCCACUCAAUUCUUGGACUUUGCUCC--CCGG ....---------------------......(((((((--(.((((((((.....)))))))).)))...))))).....(((((.((((...((........))....)))))--)))) ( -30.10) >DroYak_CAF1 60623 95 + 1 AUAU---------------------GGUUACAUCAGAU--UCCCAGUGACACGGGGUCACUGGAAAUUAACUGAUAAAAGCUGGGCGAGUAGCUCAAUUCUUGGACUUUGCUCC--CCGG ....---------------------......(((((((--(.((((((((.....)))))))).)))...))))).....(((((.((((((..((.....))..))..)))))--)))) ( -31.00) >consensus AUAU_____________________AGUUACAUCAGAU__UCCCAGUGACACGGGGUCACUGGAAAUUAACUGAUAAAAGCUGGGCGAGCAGCUCAAUUCUUGGACUUUGCUCC__CCGG ...............................(((((......((((((((.....)))))))).......))))).....((((..((((((.((((....))))..))))))...)))) (-23.72 = -24.84 + 1.13)



| Location | 14,927,246 – 14,927,361 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.68 |
| Mean single sequence MFE | -34.60 |
| Consensus MFE | -25.74 |
| Energy contribution | -26.30 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.74 |
| SVM decision value | 3.39 |
| SVM RNA-class probability | 0.999136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14927246 115 - 27905053 CCGGGGGGAGCAAAGUCCAAAGAUGGAGCUGUUCGUCCAGCUUUUAUCAGUUAAUUUCCAGUGACCCCGUGUCACUGGGAAAAUCUGAUGUAACA-AU----AUACAUACAUUCAUACAU ...(((.(((((...((((....))))..))))).)))......((((((....(((((((((((.....)))))))))))...)))))).....-..----.................. ( -35.50) >DroSim_CAF1 55921 114 - 1 CCGU--GGAGCAAAGUCCAAAAAUGGAGCUGCUCGCCCCGCUUUUAUCAGUUAAUUUCCAGUGACCCCGUGUCACUGGGAAAAUCUGAUGUAACUGAUGUAUAUACAUA----CAUACAU ..((--((.((..((((((....)))).))....)).))))...((((((....(((((((((((.....)))))))))))...))))))....((((((((....)))----))).)). ( -37.20) >DroEre_CAF1 53803 95 - 1 CCGG--GGAGCAAAGUCCAAGAAUUGAGUGGCUCGCCCAGCUUUUAUCAGUUAAUUUCCAGUGACCCCGUGUCACUGGGA--AUCUGAUGUAACU---------------------AUAU ..((--(((((...(..((.....))..).)))).)))......((((((...((((((((((((.....))))))))))--)))))))).....---------------------.... ( -33.90) >DroYak_CAF1 60623 95 - 1 CCGG--GGAGCAAAGUCCAAGAAUUGAGCUACUCGCCCAGCUUUUAUCAGUUAAUUUCCAGUGACCCCGUGUCACUGGGA--AUCUGAUGUAACC---------------------AUAU ..((--((((...((..((.....))..)).))).)))......((((((...((((((((((((.....))))))))))--)))))))).....---------------------.... ( -31.80) >consensus CCGG__GGAGCAAAGUCCAAAAAUGGAGCUGCUCGCCCAGCUUUUAUCAGUUAAUUUCCAGUGACCCCGUGUCACUGGGA__AUCUGAUGUAACU_____________________ACAU ......(((......))).......((((((......)))))).((((((.....((((((((((.....))))))))))....)))))).............................. (-25.74 = -26.30 + 0.56)



| Location | 14,927,281 – 14,927,395 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.63 |
| Mean single sequence MFE | -30.78 |
| Consensus MFE | -26.18 |
| Energy contribution | -27.10 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14927281 114 + 27905053 UCCCAGUGACACGGGGUCACUGGAAAUUAACUGAUAAAAGCUGGACGAACAGCUCCAUCUUUGGACUUUGCUCCCCCCGGCAAAUCAAUACCAAC------UACCAAAUCGUAAAUAAUU ..((((((((.....))))))))........((((....((((......))))((((....))))..(((((......)))))))))........------................... ( -28.20) >DroSec_CAF1 57283 118 + 1 UCCCAGUGACACGGGGUCACUGGAAAUUAACUGAUAAAAGCUGGGCGAGCAGCUCCAUUUUUGGACUUUGCUCC--ACGGUAAAUCAAUACGCACAACGAGUACCAAAUCUUAAAUAAUU ..((((((((.....))))))))........((((....((((.(.((((((.((((....))))..)))))))--.))))..))))................................. ( -30.50) >DroSim_CAF1 55957 116 + 1 UCCCAGUGACACGGGGUCACUGGAAAUUAACUGAUAAAAGCGGGGCGAGCAGCUCCAUUUUUGGACUUUGCUCC--ACGGCAAAUCAAUACCCACAACGAGUACCAAAG--UAAAUAAUU ..((((((((.....))))))))..................(((((.....)))))..((((((..((((((..--..))))))....(((.(.....).)))))))))--......... ( -31.10) >DroEre_CAF1 53820 112 + 1 UCCCAGUGACACGGGGUCACUGGAAAUUAACUGAUAAAAGCUGGGCGAGCCACUCAAUUCUUGGACUUUGCUCC--CCGGCAAAUCAAUACCAAC------UGCCACAUCGUAAGUAAUU ..((((((((.....))))))))........((((....((((((.((((...((........))....)))))--)))))..))))........------................... ( -31.60) >DroYak_CAF1 60640 112 + 1 UCCCAGUGACACGGGGUCACUGGAAAUUAACUGAUAAAAGCUGGGCGAGUAGCUCAAUUCUUGGACUUUGCUCC--CCGGCAAAUCAAUACCAAC------UACCACAUCGUAAAUAAUU ..((((((((.....))))))))........((((....((((((.((((((..((.....))..))..)))))--)))))..))))........------................... ( -32.50) >consensus UCCCAGUGACACGGGGUCACUGGAAAUUAACUGAUAAAAGCUGGGCGAGCAGCUCCAUUUUUGGACUUUGCUCC__CCGGCAAAUCAAUACCAAC______UACCAAAUCGUAAAUAAUU ..((((((((.....))))))))........((((....(((((..((((((.((((....))))..))))))...)))))..))))................................. (-26.18 = -27.10 + 0.92)



| Location | 14,927,281 – 14,927,395 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.63 |
| Mean single sequence MFE | -34.92 |
| Consensus MFE | -25.46 |
| Energy contribution | -26.38 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14927281 114 - 27905053 AAUUAUUUACGAUUUGGUA------GUUGGUAUUGAUUUGCCGGGGGGAGCAAAGUCCAAAGAUGGAGCUGUUCGUCCAGCUUUUAUCAGUUAAUUUCCAGUGACCCCGUGUCACUGGGA ..........(((..(((.------..(((((......)))))(((.(((((...((((....))))..))))).))).)))...))).......((((((((((.....)))))))))) ( -36.10) >DroSec_CAF1 57283 118 - 1 AAUUAUUUAAGAUUUGGUACUCGUUGUGCGUAUUGAUUUACCGU--GGAGCAAAGUCCAAAAAUGGAGCUGCUCGCCCAGCUUUUAUCAGUUAAUUUCCAGUGACCCCGUGUCACUGGGA .............((((((...((((.(((((......)))...--.(((((...((((....))))..))))))).))))...)))))).....((((((((((.....)))))))))) ( -34.90) >DroSim_CAF1 55957 116 - 1 AAUUAUUUA--CUUUGGUACUCGUUGUGGGUAUUGAUUUGCCGU--GGAGCAAAGUCCAAAAAUGGAGCUGCUCGCCCCGCUUUUAUCAGUUAAUUUCCAGUGACCCCGUGUCACUGGGA .........--..(..((((((.....))))))..)......((--((.((..((((((....)))).))....)).))))..............((((((((((.....)))))))))) ( -33.50) >DroEre_CAF1 53820 112 - 1 AAUUACUUACGAUGUGGCA------GUUGGUAUUGAUUUGCCGG--GGAGCAAAGUCCAAGAAUUGAGUGGCUCGCCCAGCUUUUAUCAGUUAAUUUCCAGUGACCCCGUGUCACUGGGA ..........((((.(((.------...((((......))))((--(((((...(..((.....))..).)))).))).)))..)))).......((((((((((.....)))))))))) ( -37.10) >DroYak_CAF1 60640 112 - 1 AAUUAUUUACGAUGUGGUA------GUUGGUAUUGAUUUGCCGG--GGAGCAAAGUCCAAGAAUUGAGCUACUCGCCCAGCUUUUAUCAGUUAAUUUCCAGUGACCCCGUGUCACUGGGA ..........((((....(------(((((((......))))((--((((...((..((.....))..)).))).)))))))..)))).......((((((((((.....)))))))))) ( -33.00) >consensus AAUUAUUUACGAUUUGGUA______GUUGGUAUUGAUUUGCCGG__GGAGCAAAGUCCAAAAAUGGAGCUGCUCGCCCAGCUUUUAUCAGUUAAUUUCCAGUGACCCCGUGUCACUGGGA ..........(((..(((..........((((......)))).....(((((...((((....))))..))))).....)))...))).......((((((((((.....)))))))))) (-25.46 = -26.38 + 0.92)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:05 2006