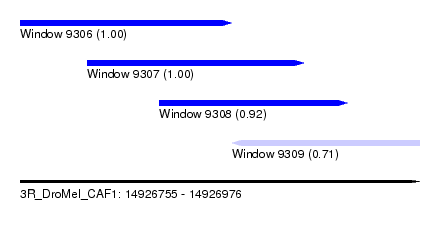
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,926,755 – 14,926,976 |
| Length | 221 |
| Max. P | 0.999703 |

| Location | 14,926,755 – 14,926,872 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.58 |
| Mean single sequence MFE | -20.08 |
| Consensus MFE | -17.86 |
| Energy contribution | -18.06 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.91 |
| SVM RNA-class probability | 0.999703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14926755 117 + 27905053 ---AAAAAUUAACGCCAAAAUAUAUAAAGCACAAACAGAGUUGAAAAAACGCAACAAAUCCGUUGAGUGUGCAAUGCACACAUGUGAUUAUAUAUCCAUACUUAUAUAAGCUAAACAAGA ---..........((..............((((......(((.....))).((((......)))).((((((...)))))).)))).(((((((........)))))))))......... ( -19.30) >DroSec_CAF1 56785 120 + 1 UAAAAAAAUUAAAUCCAAAAUAUAUAAAGCACAAGCAGAGUUGAAAAAACGCAACAAAUCCGUUGAGUGUGCAAUGCACACAUGUGAUUAUAUAUCCAUACUUAUAUAAGCUAAACAAGA ...................((((((((..((((......(((.....))).((((......)))).((((((...)))))).)))).))))))))......................... ( -19.20) >DroSim_CAF1 55418 120 + 1 UAAAAAAAUUAACGCCAAAAUAUAUAAAGCACAAGCAGAGUUGAAAAAACGCAACAAAUCCGUUGAGUGUGCAAUGCACACAUGUGAUUAUAUAUCCAUACUUAUAUAAGCUAAACAAGA ........((((((..............((....))...((((........)))).....))))))((((((...)))))).(((..(((((((........))))))).....)))... ( -21.20) >DroEre_CAF1 53331 113 + 1 -----AAAUUAACGCCAAAAUA--AGAAACACAAACAAAGGUGAAAAAACGCAACAAAUCUGUUGAGUGUGCAAUGCACACAUGUGAUUAUAUAUCCAUACUUAUAUAAGCUGUACAAGA -----.......((((......--...............))))........(((((....))))).((((((...)))))).((((.(((((((........)))))))....))))... ( -20.60) >DroYak_CAF1 60151 113 + 1 -----AAAUUAACGCCAAAAUA--AAAAACACAAACAAAGGUGAAAAAACGCAACAAAUCUGUUGAGUGUGCAAUGCACACAUGUGAUUAUAUAUCCAUACUUAUAUAAGCUAUACAAGA -----........((.......--.....((((..................(((((....))))).((((((...)))))).)))).(((((((........)))))))))......... ( -20.10) >consensus ___AAAAAUUAACGCCAAAAUAUAUAAAGCACAAACAGAGUUGAAAAAACGCAACAAAUCCGUUGAGUGUGCAAUGCACACAUGUGAUUAUAUAUCCAUACUUAUAUAAGCUAAACAAGA .............((..............((((..................((((......)))).((((((...)))))).)))).(((((((........)))))))))......... (-17.86 = -18.06 + 0.20)



| Location | 14,926,792 – 14,926,912 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -24.92 |
| Consensus MFE | -23.00 |
| Energy contribution | -23.20 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.995858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14926792 120 + 27905053 UUGAAAAAACGCAACAAAUCCGUUGAGUGUGCAAUGCACACAUGUGAUUAUAUAUCCAUACUUAUAUAAGCUAAACAAGACAUUGUCUGGGGCAUUAAAAACGCAUAACAAUUAAUUGCU ..........((((..(((..((((.((((((...)))))).((((.(((((((........)))))))(((...((.(((...))))).)))........)))))))).)))..)))). ( -25.20) >DroSec_CAF1 56825 120 + 1 UUGAAAAAACGCAACAAAUCCGUUGAGUGUGCAAUGCACACAUGUGAUUAUAUAUCCAUACUUAUAUAAGCUAAACAAGACAUUGUCUGGGGCAUUAAAAACGCAUAACAAUUAAUUGUU ............(((((....((((.((((((...))))))(((((.(((((((........)))))))(((...((.(((...))))).)))........)))))..))))...))))) ( -24.60) >DroSim_CAF1 55458 120 + 1 UUGAAAAAACGCAACAAAUCCGUUGAGUGUGCAAUGCACACAUGUGAUUAUAUAUCCAUACUUAUAUAAGCUAAACAAGACAUUGUCUGGGGCAUUAAAAACGCAUAACAAUUAAUUGUU ............(((((....((((.((((((...))))))(((((.(((((((........)))))))(((...((.(((...))))).)))........)))))..))))...))))) ( -24.60) >DroEre_CAF1 53364 120 + 1 GUGAAAAAACGCAACAAAUCUGUUGAGUGUGCAAUGCACACAUGUGAUUAUAUAUCCAUACUUAUAUAAGCUGUACAAGACAUUGUCUGGGGCAUUAAAAAAGCAUAACAAUUAAUUGCU ..........((((..(((.(((((.((((((...)))))).(((..(((((((........)))))))(((.(((((....))))..).))).........)))))))))))..)))). ( -23.60) >DroYak_CAF1 60184 120 + 1 GUGAAAAAACGCAACAAAUCUGUUGAGUGUGCAAUGCACACAUGUGAUUAUAUAUCCAUACUUAUAUAAGCUAUACAAGACAUUGUCUGGGGCAUUAAAAACGCAUAACAAUUAAUUGUU (((......)))(((((....((((.((((((...))))))(((((.(((((((........)))))))(((...((.(((...))))).)))........)))))..))))...))))) ( -26.60) >consensus UUGAAAAAACGCAACAAAUCCGUUGAGUGUGCAAUGCACACAUGUGAUUAUAUAUCCAUACUUAUAUAAGCUAAACAAGACAUUGUCUGGGGCAUUAAAAACGCAUAACAAUUAAUUGUU ...........((((......)))).((((((...))))))(((((.(((((((........)))))))(((...((.(((...))))).)))........))))).............. (-23.00 = -23.20 + 0.20)

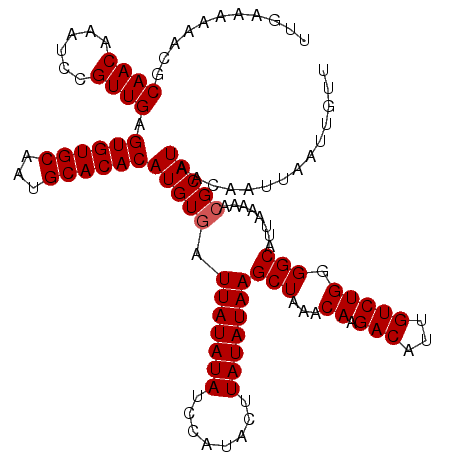

| Location | 14,926,832 – 14,926,936 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.63 |
| Mean single sequence MFE | -19.26 |
| Consensus MFE | -15.19 |
| Energy contribution | -15.11 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.917724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14926832 104 + 27905053 CAUGUGAUUAUAUAUCCAUACUUAUAUAAGCUAAACAAGACAUUGUCUGGGGCAUUAAAAACGCAUAACAAUUAAUUGCUGGAAAA----------------AAACAAUCAGCGACAGCA .(((((.(((((((........)))))))(((...((.(((...))))).)))........))))).........(((((((....----------------......)))))))..... ( -18.60) >DroSec_CAF1 56865 118 + 1 CAUGUGAUUAUAUAUCCAUACUUAUAUAAGCUAAACAAGACAUUGUCUGGGGCAUUAAAAACGCAUAACAAUUAAUUGUUGGAAAA--AAACAACAACAAAAAAACAAUCCGCGACAGCA .(((((.(((((((........)))))))(((...((.(((...))))).)))........))))).........((((((.....--...))))))..............((....)). ( -20.50) >DroSim_CAF1 55498 120 + 1 CAUGUGAUUAUAUAUCCAUACUUAUAUAAGCUAAACAAGACAUUGUCUGGGGCAUUAAAAACGCAUAACAAUUAAUUGUUGGAAAAAGAAACAACAACAAAAAAACAAUCAGCGACAGCA .(((((.(((((((........)))))))(((...((.(((...))))).)))........))))).........((((((.............))))))...........((....)). ( -19.52) >DroEre_CAF1 53404 102 + 1 CAUGUGAUUAUAUAUCCAUACUUAUAUAAGCUGUACAAGACAUUGUCUGGGGCAUUAAAAAAGCAUAACAAUUAAUUGCUGA------------------AAAAACAAUCAGCGACAGCA .......(((((((........)))))))(((((.......(((((.....((.........))...)))))....((((((------------------........))))))))))). ( -19.20) >DroYak_CAF1 60224 102 + 1 CAUGUGAUUAUAUAUCCAUACUUAUAUAAGCUAUACAAGACAUUGUCUGGGGCAUUAAAAACGCAUAACAAUUAAUUGUUGA------------------AAAAACAAUCAGCGACAGCA .(((((.(((((((........)))))))(((...((.(((...))))).)))........))))).........(((((((------------------........)))))))..... ( -18.50) >consensus CAUGUGAUUAUAUAUCCAUACUUAUAUAAGCUAAACAAGACAUUGUCUGGGGCAUUAAAAACGCAUAACAAUUAAUUGUUGGAAAA______________AAAAACAAUCAGCGACAGCA .(((((.(((((((........)))))))(((...((.(((...))))).)))........))))).........(((((((..........................)))))))..... (-15.19 = -15.11 + -0.08)



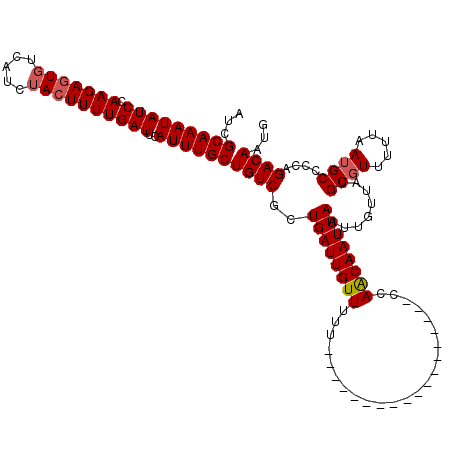
| Location | 14,926,872 – 14,926,976 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.54 |
| Mean single sequence MFE | -20.98 |
| Consensus MFE | -16.80 |
| Energy contribution | -17.30 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709462 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14926872 104 - 27905053 AUCGCAAAUAUCCAAGAGUGUCAUCUACUUUUGAUUCAUUUGCUGUCGCUGAUUGUUU----------------UUUUCCAGCAAUUAAUUGUUAUGCGUUUUUAAUGCCCCAGACAAUG ...(((((((..((((((((.....))))))))..).))))))((((((((.......----------------.....)))).............((((.....))))....))))... ( -22.10) >DroSim_CAF1 55538 120 - 1 AUCGCAAAUAUCCAAGAUUUUCAUCUACUUUUGAUUCAUUUGCUGUCGCUGAUUGUUUUUUUGUUGUUGUUUCUUUUUCCAACAAUUAAUUGUUAUGCGUUUUUAAUGCCCCAGACAAUG ...(((((((..(((((...........)))))..).))))))((((((((((.(((...((((((..(.......)..))))))..))).)))).))((.......))....))))... ( -17.30) >DroEre_CAF1 53444 102 - 1 AUCGCAAAUAUCCAAGAGUGUCAUCUACUUUUGAUUCAUUUGCUGUCGCUGAUUGUUUUU------------------UCAGCAAUUAAUUGUUAUGCUUUUUUAAUGCCCCAGACAAUG ...(((((((..((((((((.....))))))))..).))))))(((((((((........------------------))))).............((.........))....))))... ( -23.30) >DroYak_CAF1 60264 102 - 1 AUCGCAAAUAUCAAAGAGUGUCAUCUACUUUUGAUUCAUUUGCUGUCGCUGAUUGUUUUU------------------UCAACAAUUAAUUGUUAUGCGUUUUUAAUGCCCCAGACAAUG ...(((((((((((.(((((.....))))))))))..))))))((((..((((((((...------------------..))))))))........((((.....))))....))))... ( -21.20) >consensus AUCGCAAAUAUCCAAGAGUGUCAUCUACUUUUGAUUCAUUUGCUGUCGCUGAUUGUUUUU__________________CCAACAAUUAAUUGUUAUGCGUUUUUAAUGCCCCAGACAAUG ...(((((((((.(((((((.....))))))))))..))))))((((..((((((((.......................))))))))........((((.....))))....))))... (-16.80 = -17.30 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:57 2006