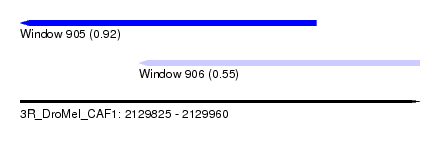
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,129,825 – 2,129,960 |
| Length | 135 |
| Max. P | 0.918600 |

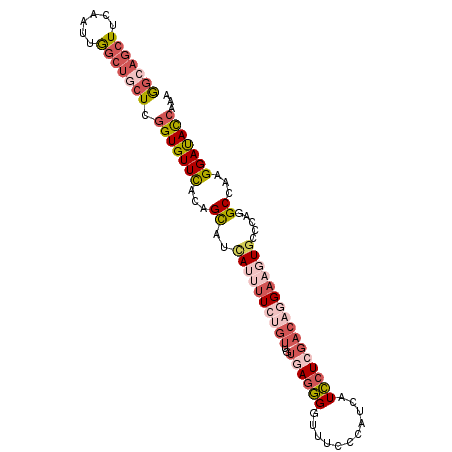
| Location | 2,129,825 – 2,129,925 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 78.93 |
| Mean single sequence MFE | -34.52 |
| Consensus MFE | -22.72 |
| Energy contribution | -25.25 |
| Covariance contribution | 2.53 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2129825 100 - 27905053 GGCAGCUUCGAUUGGCUGCUCGGUGUUCACAGCAUCAUCUUCAGUUAUGAGGGGUUUUCCAUCAUCCUCGACAGGAAGUGCCCAGGCCAAGGAUACCAAA (((((((......))))))).(((((((...((......((((....))))(((((((((.((......))..))))).))))..))...)))))))... ( -32.70) >DroSec_CAF1 273074 100 - 1 GGCAGCUUCGAUUGGCUGCUCGGUGUUCACAGCAUCAAUUUCUGUGCUGAGGGGUUUCCCAUCAUCCUCGACAGGAAGUGUCCAGGCCAAGGAUACCAAA (((((((......))))))).(((((((.((((((........)))))).(((....))).....(((.((((.....)))).)))....)))))))... ( -37.10) >DroSim_CAF1 292080 100 - 1 GGCAGCUUCAAUUGGCUGCUCGGUGUUCACAGCAUCAAUUUCUGUGCUGAGGGGUUUCCCAUCAUCCUCGACAGGAAGUGUCCAGGCCAAGGAUACCAAA (((((((......))))))).(((((((.((((((........)))))).(((....))).....(((.((((.....)))).)))....)))))))... ( -37.10) >DroEre_CAF1 276555 100 - 1 GGCAGCUUCGAUUGGCUGCUGGGUGUUCACGGCAUUAUUUUGUGUUGUGAGGGUCUUCCUAUCAUCCUCGACAGGAAGUGCCCAGGCCAAGGACACCAGC (((((((......))))))).((((((((((((((......)))))))..((((((((((.((......)).)))))).)))).......)))))))... ( -44.70) >DroYak_CAF1 277747 100 - 1 GGAUGCUUCAAUGGGCUGCUCGGUGUUCACGGCAGAAUUUUCUGUUGGUAGGGGAUUCCUAUCAUCCUCGACAGGAAGUGCCCAGGCCAAGGAUACCAAC .((.(((......)))...))(((((((..((((((....))))))(((..(((.(((((.((......)).)))))...)))..)))..)))))))... ( -35.30) >DroAna_CAF1 245927 100 - 1 AACACCUUCAAUAAAUUCCUCAGUGUUUGUGGUUUCAUCCUGAAAAGUCACAGUCGUCUCCUUAUUUUCGACAGGAAGUGUCCAGGCUAAGGACAUCAAA ...............(((((.......(((((((((.....))))..)))))((((............)))))))))((((((.......)))))).... ( -20.20) >consensus GGCAGCUUCAAUUGGCUGCUCGGUGUUCACAGCAUCAUUUUCUGUUGUGAGGGGUUUCCCAUCAUCCUCGACAGGAAGUGCCCAGGCCAAGGAUACCAAA (((((((......))))))).(((((((...((..((((((((((..((((((...........)))))))))))))))).....))...)))))))... (-22.72 = -25.25 + 2.53)

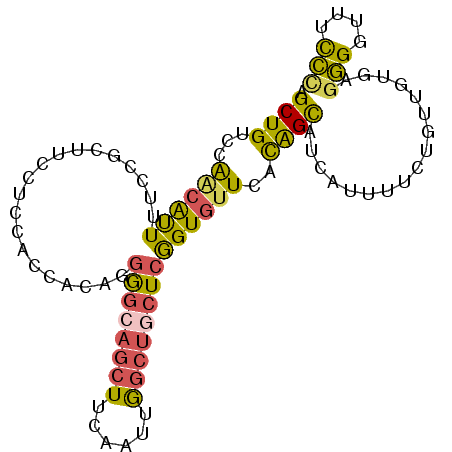
| Location | 2,129,865 – 2,129,960 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 72.84 |
| Mean single sequence MFE | -28.78 |
| Consensus MFE | -14.48 |
| Energy contribution | -14.10 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.548148 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2129865 95 - 27905053 AGCUGUCCAACAUUUUCCACUUCCUCAACCACAGGGGCAGCUUCGAUUGGCUGCUCGGUGUUCACAGCAUCAUCUUCAGUUAUGAGGGGUUUUCC .(((((..((((((.........((.......))((((((((......)))))))))))))).)))))....((((((....))))))....... ( -26.60) >DroSec_CAF1 273114 95 - 1 AGCUGUCCAACAUUUUCUGCUUCCUCCACCACAGGGGCAGCUUCGAUUGGCUGCUCGGUGUUCACAGCAUCAAUUUCUGUGCUGAGGGGUUUCCC (((...............))).((((((.(((((((((((((......))))))))(((((.....))))).....))))).)).))))...... ( -31.86) >DroSim_CAF1 292120 95 - 1 AGCUGUCCAACAUUUUCCACUUCCUCCACCACAGGGGCAGCUUCAAUUGGCUGCUCGGUGUUCACAGCAUCAAUUUCUGUGCUGAGGGGUUUCCC ......................((((((.(((((((((((((......))))))))(((((.....))))).....))))).)).))))...... ( -31.70) >DroEre_CAF1 276595 95 - 1 AGCUGUCCAACAUUUUCCGCUUCCUCCACCACCGGGGCAGCUUCGAUUGGCUGCUGGGUGUUCACGGCAUUAUUUUGUGUUGUGAGGGUCUUCCU ....(.((.....................((((.((.(((((......))))))).))))((((((((((......)))))))))))).)..... ( -29.70) >DroYak_CAF1 277787 95 - 1 AGCUGAGCAGCACUUUCUGCAUUUUCCACCACAGGGGAUGCUUCAAUGGGCUGCUCGGUGUUCACGGCAGAAUUUUCUGUUGGUAGGGGAUUCCU .((((((((((.((....(((((..(.......)..)))))......)))))))))))).....(((((((....)))))))...(((....))) ( -35.60) >DroAna_CAF1 245967 95 - 1 AGCUGACCGUGGUUCGAGGACUCUUCUAUUACUGGAACACCUUCAAUAAAUUCCUCAGUGUUUGUGGUUUCAUCCUGAAAAGUCACAGUCGUCUC ((.((((.(((((((.((((.....((((((((((...................))))))...)))).....)))))))...)))).)))).)). ( -17.21) >consensus AGCUGUCCAACAUUUUCCGCUUCCUCCACCACAGGGGCAGCUUCAAUUGGCUGCUCGGUGUUCACAGCAUCAUUUUCUGUUGUGAGGGGUUUCCC .((((...((((((....................((((((((......))))))))))))))..)))).................(((....))) (-14.48 = -14.10 + -0.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:19 2006