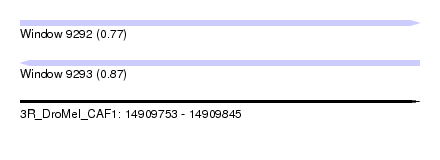
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 14,909,753 – 14,909,845 |
| Length | 92 |
| Max. P | 0.871846 |

| Location | 14,909,753 – 14,909,845 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
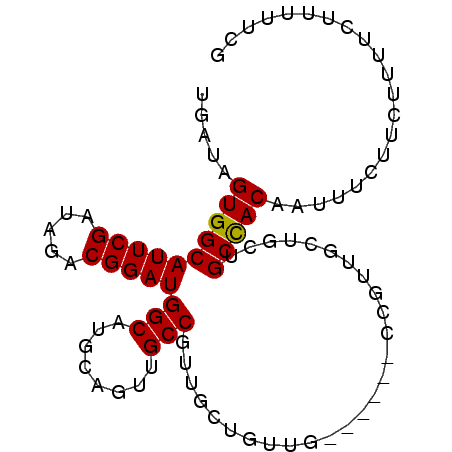
| Reading direction | forward |
| Mean pairwise identity | 82.42 |
| Mean single sequence MFE | -17.98 |
| Consensus MFE | -12.68 |
| Energy contribution | -13.08 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14909753 92 + 27905053 CGAAAAAGAAAAGAAGAAAUUGUGGCAGCAACAACAGCAAAUGCAACAGCAACGGCAACUGCAUGCCAUCCGUCUAUCGAAUGCCACUAUCA ...............((....((((((((.......))...(((....)))..((((......))))..............))))))..)). ( -18.60) >DroSec_CAF1 36979 92 + 1 CGAAAAAGAAAAGAAGAAAUUGUGGCAGCAGCAACAGCAAAUGCAACAGCAACGGCAACUGCAUGCCAUCCGUCUAUCGAAUGCCACUAUCA ...............((....((((((((((((...((...(((....)))...))...))).)))....((.....))..))))))..)). ( -20.90) >DroSim_CAF1 36935 74 + 1 CGAAAAGGAAAAGAAGAAAUUGUGGC------------------AACAGCAACGGCAACUGCAUGCCAUCCGUCUAUCGAAUGCCACUAUCA ......((....(((((....(((((------------------(...(((..(....)))).))))))...))).)).....))....... ( -15.90) >DroEre_CAF1 36385 77 + 1 CGAAAAAGAAAA---GAAAUUGUAGCAACAGCAACGG------C------AACGGCAACUACAAGCCAUCCGUCUAUCGAAUGCCACUAUCA .((.........---......((.((....)).))((------(------(..(((........)))...((.....))..))))....)). ( -13.60) >DroYak_CAF1 43354 83 + 1 CGAAAAAGAAAA---GAAAUUGUGGCAACAGCAACGG------CAACGGCAACGGCAACUGCAAGCCAUCCGUCUAUCGAAUGCCACUAUCA ............---((....((((((........((------(..((....))((....))..)))...((.....))..))))))..)). ( -20.90) >consensus CGAAAAAGAAAAGAAGAAAUUGUGGCAACAGCAACAG______CAACAGCAACGGCAACUGCAUGCCAUCCGUCUAUCGAAUGCCACUAUCA ...............((....((((((..........................(((........)))...((.....))..))))))..)). (-12.68 = -13.08 + 0.40)

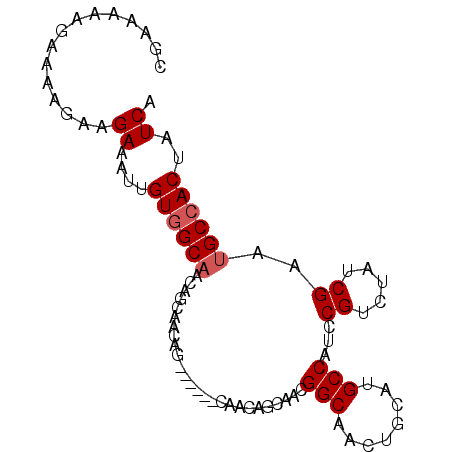
| Location | 14,909,753 – 14,909,845 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 82.42 |
| Mean single sequence MFE | -24.52 |
| Consensus MFE | -16.68 |
| Energy contribution | -16.52 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 14909753 92 - 27905053 UGAUAGUGGCAUUCGAUAGACGGAUGGCAUGCAGUUGCCGUUGCUGUUGCAUUUGCUGUUGUUGCUGCCACAAUUUCUUCUUUUCUUUUUCG .....((((((..((((((.((((((.((.(((((.......)))))))))))))))))))....))))))..................... ( -27.80) >DroSec_CAF1 36979 92 - 1 UGAUAGUGGCAUUCGAUAGACGGAUGGCAUGCAGUUGCCGUUGCUGUUGCAUUUGCUGUUGCUGCUGCCACAAUUUCUUCUUUUCUUUUUCG .....((((((((((.....)))))((((.((((..((...(((....)))...))..)))))))))))))..................... ( -27.40) >DroSim_CAF1 36935 74 - 1 UGAUAGUGGCAUUCGAUAGACGGAUGGCAUGCAGUUGCCGUUGCUGUU------------------GCCACAAUUUCUUCUUUUCCUUUUCG .....((((((....((((...(((((((......))))))).)))))------------------)))))..................... ( -21.20) >DroEre_CAF1 36385 77 - 1 UGAUAGUGGCAUUCGAUAGACGGAUGGCUUGUAGUUGCCGUU------G------CCGUUGCUGUUGCUACAAUUUC---UUUUCUUUUUCG .....((((((..((...((((((((((........))))).------.------)))))..)).))))))......---............ ( -21.30) >DroYak_CAF1 43354 83 - 1 UGAUAGUGGCAUUCGAUAGACGGAUGGCUUGCAGUUGCCGUUGCCGUUG------CCGUUGCUGUUGCCACAAUUUC---UUUUCUUUUUCG .....((((((..((((.((((((((((........)))))..))))).------..))))....))))))......---............ ( -24.90) >consensus UGAUAGUGGCAUUCGAUAGACGGAUGGCAUGCAGUUGCCGUUGCUGUUG______CCGUUGCUGCUGCCACAAUUUCUUCUUUUCUUUUUCG .....((((((((((.....)))))(((........)))...........................)))))..................... (-16.68 = -16.52 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:41 2006